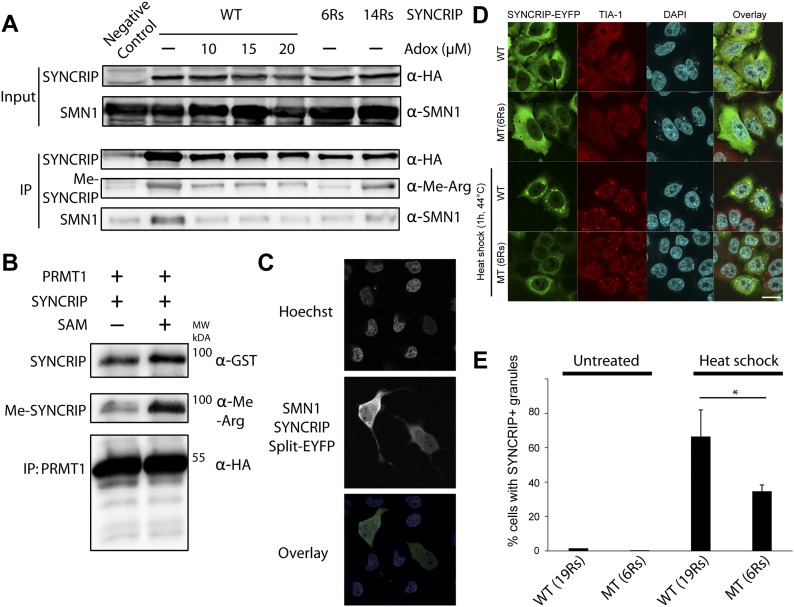
Figure 3. Characterization of SYNCRIP-SMN1 methylation–dependent interaction.
(A) Methylation dependency of SMN1-SYNCRIP interaction. Immunoprecipitation (IP) of SYNCRIP-HA from HEK293T cells expressing either WT SYNCRIP in the presence or absence of Adox methylation inhibitor, or SYNCRIP containing a reduced number of arginines. (B) SYNCRIP binding and subsequent in vitro methylation by HEK293T cell–produced PRMT1 in the presence of cofactor SAM. (C) Confocal live micrographs of HeLa cells expressing SYNCRIP and SMN1 each tagged with a part of the EYFP fluorophore, Hoescht used to visualize the nucleus. EYFP reconstitutes upon SYNCRIP-SMN1 binding, allowing subsequent visualization. Scale bar indicates 10 μm. (D) Under nonstress conditions, WT SYNCRIP and a mutant containing only six arginines show the same general subcellular localization pattern. Upon heat shock, stress granules (stained with TIA-1 stress granule protein marker) form in both WT and mutant expressing cells. WT SYNCRIP is recruited to stress granules significantly more efficiently than the six arginine SYNCRIP mutant. Scale bar, 20 μm. (E) Quantification of wild and mutant SYNCRIP recruitment to stress granules. Number of cells with and without SYNCRIP-positive stress granules was counted (n = 100) in four independent experiments. Graph shows mean of four independent experiments, error bars indicate standard deviation. Two sided Mann–Whitney–Wilcoxon test used to determine statistical significance. *P < 0.05.