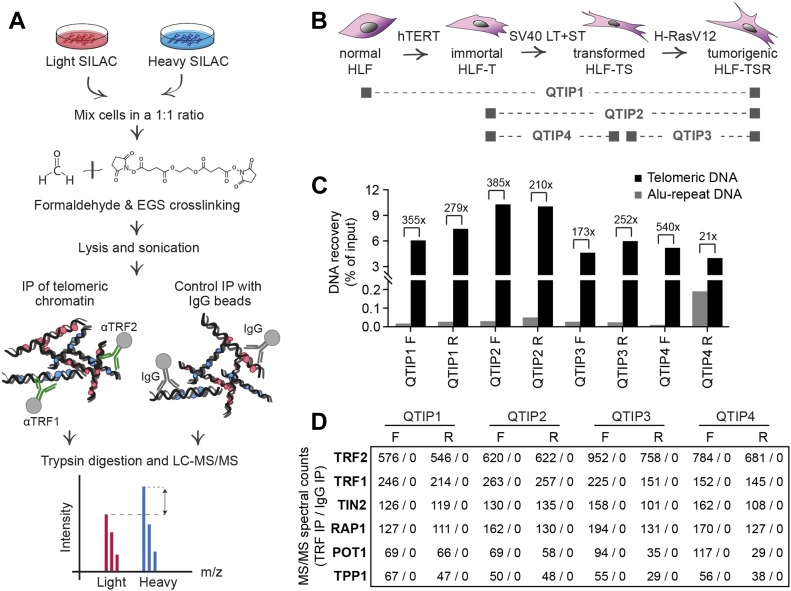
Figure 1. QTIP method was used to characterize changes in telomere protein composition during cellular transformation.
(A) Workflow of QTIP. (B) Schematic of the cell line model and overview of the four pairwise QTIP experiments. HLFs were serially transduced with retroviral vectors expressing hTERT, the SV40 large T (LT) and small t (ST) antigens, and H-RasV12. (C) Quantification of precipitated DNA in QTIPs, based on a dot blot hybridized with a specific telomeric probe and a control Alu repeat probe (Fig S3). To determine IP efficiency, the amounts of telomeric DNA in QTIP eluates were quantified and compared with the telomeric DNA in inputs. Fold enrichment of precipitated telomeric DNA compared with precipitated Alu repeat DNA is used as an indicator of IP specificity. Plotted are values from the forward (F) and reverse (R) TRF IP replicates. (D) Enrichment of shelterin subunits in QTIPs. The mean of spectral counts of two replicates is indicated.