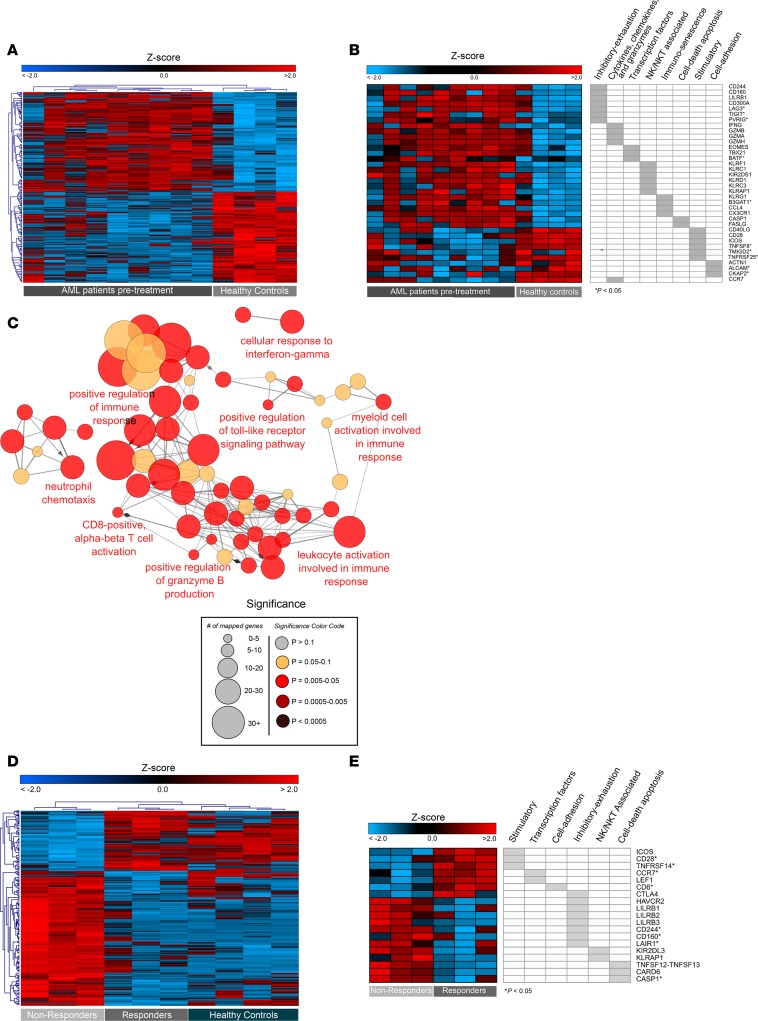
Figure 4. Differential gene expression underscoring CD8+ T cell dysfunction in AML at diagnosis and in nonresponders following induction chemotherapy.
(A–C) Gene expression analysis of PB CD8+ T cells from AML patients at diagnosis (n = 9) and age-matched HCs (n = 4). (A) Hierarchical clustering of 453 DEGs between patients and HC (log2FC > 1 and < –1, and P < 0.01). Log2 intensities are gene-wise Z transformed across all samples and visualized as heatmap. Every row represents a gene, and every column a patient sample. Red indicates an increase over the mean (Z > 0) and blue a decrease (Z < 0). (B) Heatmap of select DEGs grouped into key biological categories (log2FC > 1 and < –1; P < 0.01; *P < 0.05). (C) GO terms and KEGG/BioCarta pathways (P < 0.01) were functionally grouped into networks and interconnected based on the number of shared genes (κ score > 0.4) using ClueGO. The size of the nodes correlates with the number of mapped genes per term and color correlates with P values. (D and E) Gene expression analysis of PB CD8+ T cells from AML patients after induction chemotherapy (n = 6), 3 with complete remission (CR) and 3 nonresponders (NRs), and age-matched HCs (n = 4). (D) Hierarchical clustering of 351 DEGs from CR versus NR patients after treatment (log2FC > 1 or < –1; and P < 0.01). Expression levels for the 351 genes were queried from 4 HCs and included into the hierarchical clustering of the heatmap. Log2 intensities are gene-wise Z transformed across all samples. Every row represents a gene, and every column a patient sample. Red indicates an increase over the mean (Z > 0) and blue a decrease (Z < 0). (E) Heatmap of representative DEGs (CR versus NR) grouped into key biological categories (log2FC > 1 or < –1; P < 0.01; *P < 0.05).