Extended Data Fig. 7 |. Microbial community structures are similar between Tet2−/− mice and littermate controls.
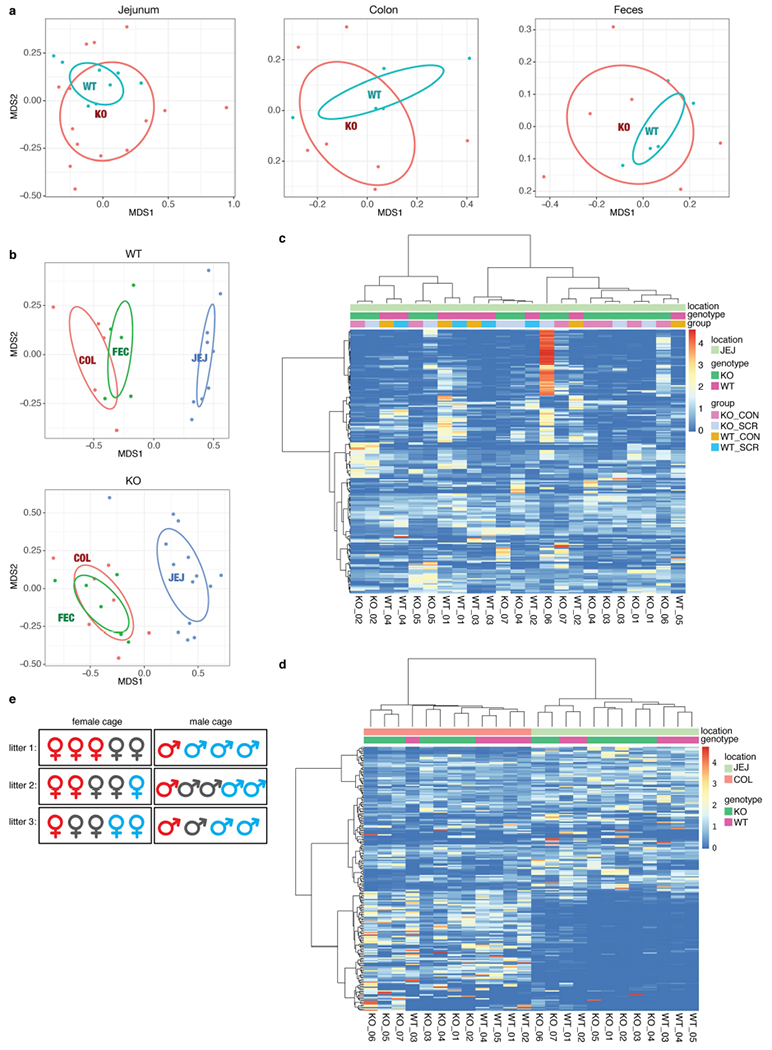
a, b, Non-metric multidimensional scaling of samples based on their oligotype profiles (with Bray–Curtis distance). a, Microbial community structures within the same body site—that is, jejunum (left, n = 14 (Tet2 (KO)) or 9 mice (Tet2+/+(WT))), colon (middle, n = 7 (KO) or 5 mice (WT)) and faeces (right, n = 6 (KO) or 5 mice (WT))—do not differ significantly across genotypes based on envfit test with 999 permutations (P > 0.05, two-tailed). b, Microbial community structures within the same genotype—WT (top, n = 5 (colonic sample (COL) or faecal sample (FEC)) or 9 mice (jejunal sample (JEJ)) and KO (bottom, n = 6 (FEC), 7 (COL) or 14 mice (JEJ))—significantly differ across body sites based on the same test (P < 0.001). c, d, Heat-map displays of oligotypes across samples. Clustering dendrograms are computed with Bray–Curtis distance and average linkage algorithm using the oligotype profiles. The intensity of colours indicates the per cent abundance of a given oligotype (rows) in a given sample (columns). c, Comparison of genotypes for samples collected from a single body site. d, Comparison of genotypes for samples collected across body sites. e, Co-housing does not affect the development of PMP in Tet2f/fVavcre mice. Representative cage configurations of 3 litters from Tet2f/fVavcre mice with PMP that are over 20 weeks old (red), symptom-free Tet2f/fVavcre mice (grey), and littermate controls (blue). CON, luminal content; SCR, scraping samples.
