Fig. 3 |. Microbial signals are required for PMP in Tet2−/− mice.
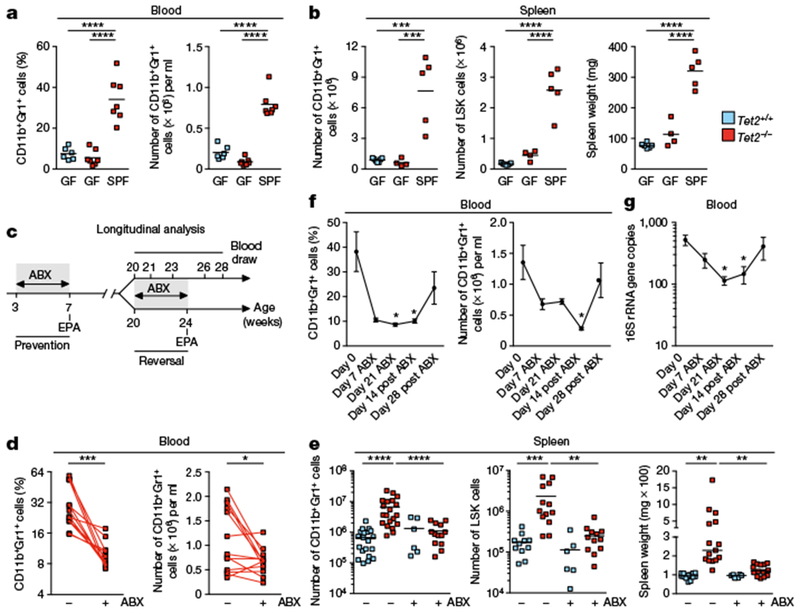
a, b, Germ-free (GF), SPF-housed (SPF) Tet2−/− mice that are over 40 weeks old, and littermate controls, analysed for percentage (a, left) and numbers (a, right) of CD11b+Gr1+ myeloid cells (n = 6 (GF Tet2+/+), 7 (GF Tet2−/−) or 7 (SPF Tet2−/−) mice), and numbers of CD11b+Gr1+ myeloid cells (b, left), LSK cells (b, middle), and spleen weight (b, right), (n = 7 (GF Tet2+/+), 4 (GF Tet2−/−) or 5 (SPF Tet2−/−) mice). c, Schematic for antibiotics (ABX) treatment regimen. Longitudinal analysis described in f, g and Extended Data Fig. 8d; prevention described in Extended Data Fig. 8c; and reversal described in d, e and Extended Data Fig. 9a–j. EPA, end point analysis. d, e, Tet2−/− mice with PMP that are over 20 weeks old, and littermate controls, analysed for percentage (d, left) and numbers (d, right) of CD11b+Gr1+ myeloid cells (n = 13 mice). Lines connect values obtained from the same mouse sampled before (−) and after (+) ABX treatment. Two-tailed paired t-test. e, Numbers of CD11b+Gr1+ myeloid cells (left; n = 20 (Tet2+/+, no ABX treatment), 20 (Tet2−/−, no ABX treatment), 6 (Tet2+/+, with ABX treatment) and 13 (Tet2−/−, with ABX treatment) mice), LSK cells (middle; n = 11 (Tet2−/−, no ABX treatment), 13 (Tet2−/−, no ABX treatment), 6 (Tet2−/− with ABX treatment) and 12 (Tet2−/−, with ABX treatment) mice) and spleen weight (right; n = 14 (Tet2+/+, no ABX treatment), 15 (Tet2−/−, no ABX treatment), 6 (Tet2+/+, with ABX treatment) and 14 (Tet2−/−, with ABX treatment) mice). f, g, Tet2−/− mice analysed for frequency (f, left) and number (f, right) of CD11b+Gr1+ myeloid cells and (g) 16S gene copies before, during and after ABX treatment (n = 7 mice). Mean ± s.e.m., repeated measures one-way ANOVA, Sidak’s post hoc test. a, b, e, Centre is mean, one-way ANOVA, Sidak’s post hoc test. Data are representative of at least two independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
