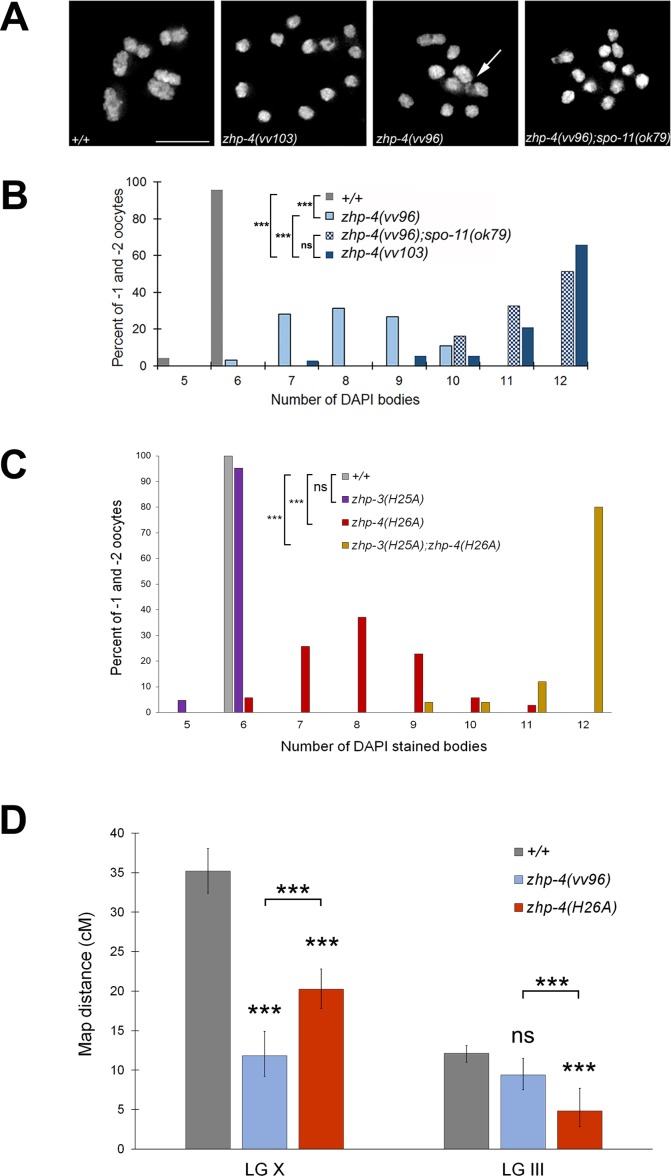
Fig 3. zhp-4 mutants are compromised for crossing over.
(A) Representative full projections of diakinesis nuclei of the indicated genotypes stained with DAPI. In comparison to the six bivalents observed in wild types, zhp-4(vv103) mutants contain no bivalents, indicating a loss of crossover formation. In contrast, zhp-4(vv96) mutants nuclei contain a mixture of structures, including univalents, anomalous bivalents ‘tethered’ by chromatin (white arrow), and bivalents. zhp-4(vv96);spo-11(ok79) nuclei contain mostly univalents, indicating that the bivalents displayed by zhp-4(vv96) are recombination initiation dependent. (B) Histogram showing distribution of the number of DAPI bodies in diakinesis nuclei in the -1 and -2 oocytes for the indicated genotypes. Nuclei scored: +/+ n = 97, zhp-4(vv103) n = 38, zhp-4(vv96) n = 64, zhp-4(vv96);spo-11(ok79) n = 30. Using Kruskal-Wallis and Dunn’s post test, all pairwise comparison results are significantly different (*** p<0.001) except for the comparison between zhp-4(vv103) and zhp-4(vv96);spo-11(ok79) nuclei (ns p>0.05). (C) Bar graph showing quantification of the numbers of DAPI-stained bodies in diakinesis nuclei at the -1 and -2 positions in germlines from animals of the indicated genotype. Most zhp-3(H25A) nuclei have 6 DAPI bodies and are not significantly different from wild types while the increase observed in zhp-4(H26A) mutants is significantly different from both. zhp-3(H25A);zhp-4(H26A) has on average 12 DAPI bodies indicating a severe loss of function (significantly different from all other genotypes assessed). Statistical significance assessed by Kruskal-Wallis test and post Dunn’s test: ns = not significant, p>0.05, *** p<0.001. Nuclei scored: +/+ n = 20, zhp-3(H25A) n = 21, zhp-4(H26A) n = 35, zhp-3(H25A);zhp-4(H26A) n = 25. (D) Bar graph showing the frequency of genetic recombination in two large genetic intervals in wild types and zhp-4 mutants (detailed in Materials and Methods). Multiple pairwise coparisons indicate that wild types and zhp-4 mutants are all significantly different from one another in the X chromosome dpy-3(e27) unc-3(e151) interval. In the dpy-18(e364) unc-25(e156) interval on chromosome III, no statistical difference was observed between wild types and zhp-4(vv96) mutants, but zhp-4(H26A) mutants are statistically different from both wild types and zhp-4(vv96) mutants. Data are represented as recombination frequency results ± 95% confidence interval and statistical analysis was conducted using Chi-squared test and Bonferroni corrections on the raw count of phenotypes (ns p>0.05, *** p<0.001).