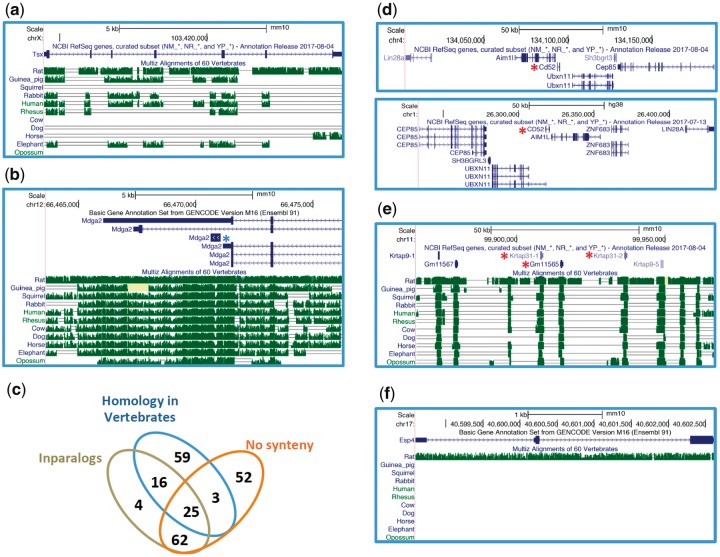
Fig. 2.
—Examples of de novo and “de nono” rodent genes visualized through the USCS Genome Browser. (a) An intergenic de novo gene with relatively low synteny conservation across several nonrodent mammals. (b) A de novo gene (blue asterisk) overlapping with the 3′UTR of an older gene. Notice that the gene symbol is the same for the two genes, which share no coding or protein similarity. (c) Summary of “de nono” genes features. (d) A “de nono” gene (Cd52, red asterisk) with conserved flanking genes in mouse (top) and human (bottom). (e) A tandem array of keratin-associated genes including three “de nono” genes (red asterisks). (f) A “de nono” gene with no synteny conservation beyond rat. Coding exons, UTRs and introns are shown as thick blue bars, thin blue bars and lines with arrows, respectively. When annotated, alternative transcripts are shown. The conservation track (green bars and single or double lines) represents the level of sequence identity between the highlighted mouse genomic region and its orthologous regions in other mammals, based on MultiZ alignments of 60 vertebrate genomes. The height of the green bars is proportional to the level of nucleotide sequence identity. The single line indicates no bases in the aligned species due to indels, whereas the double line shows regions with one or more unalignable bases. The pale yellow coloring indicates regions with Ns. The lack of any feature in the conservation track designates a region with no alignable bases and thus a complete lack of synteny conservation.