Abstract
Specific RNA structures control numerous metabolic processes that impact human health, and yet efforts to target RNA structures de-novo have been limited. In eukaryotes, the self-splicing group II intron is a mitochondrial RNA tertiary structure that is absent in vertebrates but essential for respiration in plants, fungi and yeast. Here we show that this RNA can be targeted through a process of high-throughput in vitro screening, SAR and lead optimization, resulting in high affinity compounds that specifically inhibit group IIB intron splicing in vitro and in vivo and lack toxicity in human cells. The compounds are potent growth inhibitors of the pathogen Candida parapsilosis, displaying antifungal activity comparable with Amphotericin B. These studies demonstrate that RNA tertiary structures can be successfully targeted de-novo, resulting in pharmacologically valuable compounds.
Introduction.
It is becoming increasingly clear that large, highly structured RNA molecules are essential for most metabolic functions. In theory, RNA molecules present novel targets for drug discovery, but with the exception of bacterial riboswitch inhibitors 1, small molecule inhibitors of RNA tertiary structures have not been successfully identified using the high-throughput screening and classical medicinal chemistry campaigns that have yielded most protein inhibitors. There are, to our knowledge, no new classes of antimicrobial compounds that target RNA, and none that are designed to target eukaryotic pathogens. While most inhibitors operate at the protein level, small molecules that target specific RNA structures can potentially modulate gene expression through a diversity of mechanisms, representing a powerful orthogonal strategy for the treatment of disease.
Previous studies on bacterial riboswitches have demonstrated that folded RNA molecules often bind small molecules such as metabolites with high affinity, and these ligands have been optimized in efforts to develop new antibiotics1–7. Likewise, group I introns and HIV-1 TAR RNA are known to bind small molecules8–11. Indeed, antibacterial compounds have long been known to target ribosomal RNA12,13 and, given the availability of high resolution structural data, these have been the subject of continuing optimization14,15. There has also been a growing interest in understanding structural and physicochemical properties of small molecules that selectively bind RNA motifs16,17. While these studies provide important precedents, they were conducted on RNA molecules with known small molecule ligands. De-novo RNA targeting efforts by HTS and other methods have been limited, and they have focused primarily on small RNA secondary structural elements, such as junctions, miRNAs, RNA hairpins from triplet repeat diseases and stem-loops in viral RNA genomes11,18–24. It would be ideal to identify small molecules that selectively bind RNA tertiary structures, as these more complex RNA motifs provide a route to specific molecular recognition and they are often found in biomolecules of high medical relevance.
To accomplish this goal, we have focused on RNA targets within pathogenic yeasts, as these organisms have become increasingly problematic, particularly for patients with compromised immune systems, such as recipients of implanted devices, neonatal patients, and cancer patients 25. For example, there has been a marked increase in pathologies associated with non-albican strains, particularly Candida parapsilosis 26. The availability of potent antifungals that lack toxicity in mammals is a major unmet medical need, and of value for industrial and agricultural applications. The development of new antifungals is difficult because, as eukaryotes, fungi and yeast cells have enzymes and biochemical pathways that are similar to those of humans. However, fungal RNA metabolism differs significantly, thereby providing a potential route toward new therapeutics 27
To meet these challenges, we set out to identify small molecule inhibitors of group II introns, which are large self-splicing ribozymes that are found in the mitochondrial genomes of plants, fungi, and yeast, but are not present in mammals. These autocatalytic RNA molecules adopt an elaborate tertiary structure that has been crystallographically characterized and which contains an active-site for RNA cleavage and ligation as well as solvent-accessible pockets for potential inhibitor binding28,29. In yeasts such as Saccharomyces cerevisiae (S. cerevisiae) and the pathogen Candida parapsilosis (C. parapsilosis), group II introns are found within genes that are essential for respiration, such as cytochrome oxidase subunit genes of the mitochondria30,31. Importantly, respiration is essential for pathogenic yeast to differentiate into biofilms, which colonize medical implant surfaces, are relatively resistant to antifungals, and contribute to pathogenic virulence 27,31,32. Thus, based on the complexity of their structures, and their essential role in fungal metabolism, group II introns represent outstanding targets for the development of highly specific antifungal agents.
Results.
High-throughput screening.
To identify group II intron splicing inhibitors, we developed a sensitive, high-throughput fluorescent assay for monitoring ribozyme activity of the well-characterized ai5γ group II intron from S. cerevisiae (Supplementary Fig. 1a, Supplementary Fig. 2, Supplementary Table 1, Fig. 1a). The self-splicing ai5γ group II intron was transformed into a multiple-turnover ribozyme by removing the flanking exons and consolidating catalytic domains to create the D135 ribozyme, which efficiently and specifically catalyzes cleavage of RNA “substrate” oligonucleotides that contain sequences of the original 5’-splice site 33. The RNA substrate for the screening assay was an oligonucleotide containing the last 17 nucleotides of the 5’-exon (which encompasses the “intron binding sequences” that base-pair with intron Domain 1) and the first two nucleotides of the intron (Substrate 17/2 DL, Supplementary Fig. 2a). A fluorophore and a fluorescence quencher were conjugated at positions on opposite sides of the scissile linkage so that fluorescence is observed when the oligonucleotide substrate is cleaved by the ribozyme (Supplementary Fig. 2b-c).
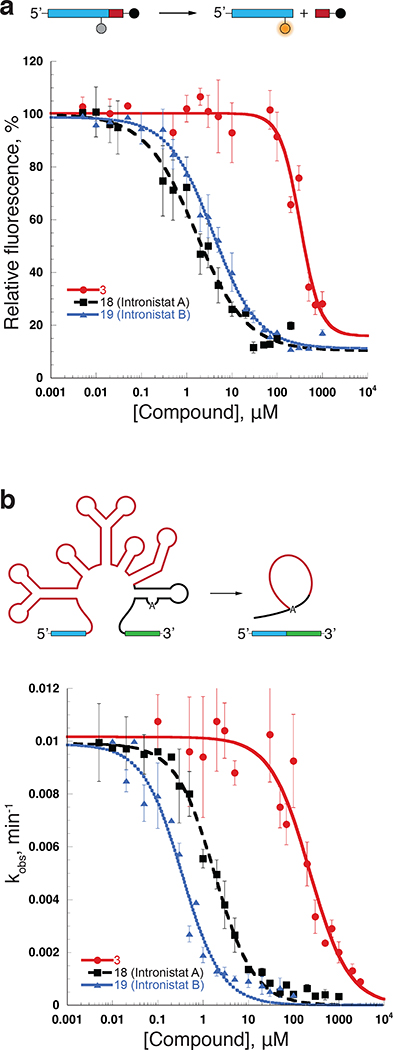
Figure 1.
The assays for compound development. (a) Top, the schematic of the fluorescent assay used for HTS and for the IC50 determination. Bottom, representative IC50 values (Supplementary Table 2) were obtained by plotting relative fluorescence vs the inhibitor concentration. Data represent average of n=3 independent experiments, error bars are s.e.m. (b) Top, the schematic of the self-splicing assay used to determine Ki values for the group II intron splicing inhibitors. Bottom, representative Ki values (Fig. 2, Supplementary Table 2) were determined by plotting the rate constants (kobs values, Supplementary Fig. 3) vs. inhibitor concentration. Data represent average of n=3 independent experiments for 18 (Intronistat A) and 19 (Intronistat B) and n=4 independent experiments for 3. Error bars are s.e.m.
Using this assay for ribozyme activity, we screened a curated library of 10,000 compounds (see Methods), identifying 16 reproducible hits, some of which shared common structural elements that suggested a shared mechanism of action (Supplementary Fig. 2d). We then analyzed a series of commercially available derivatives of the major hits in order to identify more suitable scaffolds for further optimization. The most potent scaffold identified during this phase of the study was compound 1, which exhibited an IC50 of 2 μM (Supplementary table 2). It was used as a starting point for the design of additional compounds that were used to define structure activity relationships (SAR) and to optimize potency (Supplementary table 2).
In vitro SAR and optimization of potency.
In order to carry out the SAR studies in vitro, the primary fluorimetric assay was complemented by a robust secondary radioanalytic self-splicing assay of the precursor RNA containing the full length ai5γ intron and short exons, which enabled the determination of Ki values for all compounds of interest (Fig. 1b, Supplementary Fig. 3a-c, Supplementary table 2).
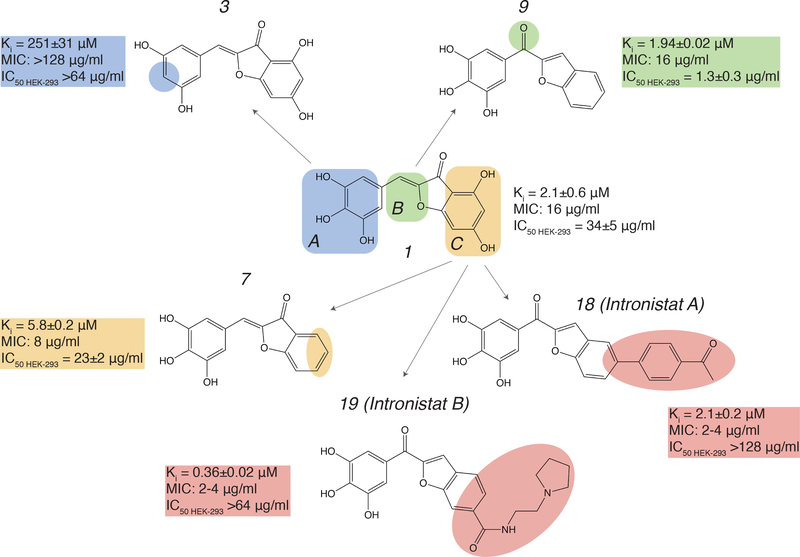
As part of an initial strategy to optimize the early leads, a series of compounds was selected to determine the critical structural components required for activity: the pharmacophore. Three regions of compound 1 (A, B and C) were defined and substituents in these sections of the molecule were evaluated for their effect on inhibition (Fig. 2). In region A, replacing any of the hydroxyl groups with hydrogen or methoxy or boronic acid substituents (compounds 2-5, Supplementary table 2) all resulted in nearly complete loss of activity. Notably, when the trihydroxyl was replaced with a dihydroxyl catechol moiety, the molecule was inactivated both in vitro and in vivo (2, Supplementary table 2), indicating that presence of the catechol motif by itself cannot explain reactivity of the group II intron inhibitors, unlike certain classes of promiscuous molecules that are collectively classified as “PAINS” compounds.
Figure 2.
Summary of SAR studies on compound 1. In vitro splicing inhibition constants (Ki) for the ai5γ intron, MIC values for C. parapsilosis and IC50 values for cytotoxicity in HEK-293 cells are shown next to each molecule. Color-coding is used to highlight functional regions of the lead compound. Changes in Region A of compound 1 are shown in blue, changes in Region B are shown in green, and changes in Region C are shown in yellow. Changes that were introduced for optimization of inhibitory activity are highlighted in pink.
In contrast, each of the hydroxyl groups in region C could be removed or replaced with halogen atoms without significant loss of function (compounds 6-8, Fig. 2, Supplementary table 2). To minimize any risk of reactivity from the α,β-unsaturated ketone, we replaced the 2-benzylidenebenzofuran-3(2H)-one moiety with a more chemically and metabolically stable benzofuran-2-yl(phenyl)methanone (region B). This more drug-like molecular template resulted in a 2-fold increase in in vitro potency compared with the parental molecule 7 (compound 9, Fig. 2, Supplementary table 2). Other attempts to introduce changes in this region (for example, amide or thiazole derivatives) resulted in a substantial loss of activity (compounds 10 and 11, Supplementary table 2). The fact that potency was sensitive to certain modifications in Region B, and in distal parts of Region C (Supplementary table 2), indicates that inhibitory activity is not solely attributable to functional groups in Region A.
With the new lead structure (compound 9) in hand, additional substituents were added to the benzofuran moiety to further develop the structure-activity relationship for the series. Introduction of a wide variety of substituents, including aryl, heteroaryl, amino or halogens at the 5’ or 6’-positions were all generally tolerated (compounds 8,12,13,14-19, Fig. 2, Supplementary Fig. 3c, Supplementary table 2). Notably, our data suggest that adding large substituents or positively charged residues to region C can increase inhibitory activity (lower the Ki) of the respective compounds (Fig. 2, Supplementary table 2). Examples of such effects are observed for compounds 13, 14 and 19 (Fig. 1b, Fig. 2, Supplementary Fig. 3c, Supplementary table 2).
Reversibility of inhibitor binding to the intron was established using pulse-chase dilution experiments (Supplementary Fig. 3d, see Methods). The clear reactivity patterns evident from this SAR analysis, together with facile reversibility of compound binding and inhibition are consistent with selective behavior.
Inhibition of group II splicing in S. cerevisiae in vivo.
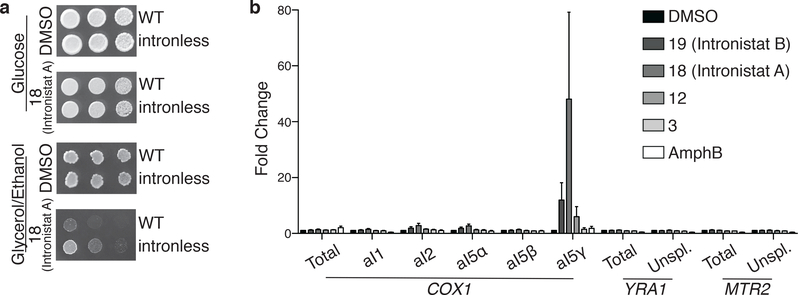
It was important to determine whether compounds that disrupt splicing of the ai5γ intron in vitro can also inhibit splicing of the intron in vivo. The ai5γ group II intron interrupts a gene encoding the first subunit of the cytochrome oxidase (COX1) in S. cerevisiae, and respiratory-deficient petite colonies are therefore formed when ai5γ intron splicing is disrupted. These colonies cannot grow on non-fermentable carbon sources like glycerol, but they exhibit growth on glucose 30. To determine whether our inhibitory compounds might induce a respiratory defect, we compared the growth of S. cerevisiae in both YPD and YPGE media in the presence of a panel of inhibitors. We found that growth of S. cerevisiae in the presence of inhibitory small molecules is inhibited in glycerol/ethanol medium (YPGE), but not in glucose medium (YPD) (Supplementary table 3), which is consistent with the expected respiratory defect.
To analyze whether the growth defect observed in the presence of inhibitor is specifically due to defective splicing of the ai5γ intron, we tested whether the compounds inhibited growth of an “intronless strain”, which contains an intact COX1 subunit gene from which the intron has been removed, thereby obviating the requirement for splicing 34. We observed a clear difference in growth in between the intronless strain and the wild-type strain in the presence of compound 18, revealing that the intronless strain is much more resistant to compound 18 in YPGE medium (Fig. 3a, Supplementary Fig. 4a). This behavior is consistent with specific inhibition of the ai5γ intron in vivo. That said, growth of the intronless strain is somewhat slower in the presence of inhibitor, suggesting that 18 may cause certain off-target effects.
Figure 3.
Active compounds selectively inhibit group II splicing in vivo. (a) Comparison of wild-type (WT) and intronless S. cerevisiae strain growth in the presence of 18 (Intronistat A). From left to right, approximately 22,500, 4,500, and 900 cells of the indicated strains were plated on either YP 2% glucose or YP 3% glycerol 3% ethanol media containing DMSO vehicle or compound 18 (Intronistat A) at 128μg/mL for growth at 30°C. (b) Active compounds inhibit splicing of COX1 aI5γ group II intron in vivo. S. cerevisiae were grown in the presence of DMSO vehicle, active compounds 19 (Intronistat B), 18 (Intronistat A), and 12, inactive compound (3), or amphotericin B (AmphB). Relative levels of total and unspliced levels of COX1 indicated by qRT-PCR quantification of amplicons covering the eighth exon (total) or the intron-exon junction from the group IIA introns aI1 and aI2, the group I introns aI5α and aI5β, or the group IIB intron aI5γ. Levels of total and unspliced nuclear-encoded YRA1 and MTR2 indicated by quantification of amplicons covering an exon (total) or intron-exon junction of the splicesomal targeted intron. Mean values and s.e.m. from n=4 independent experiments are shown with ACT1 as a standard.
To directly monitor the effect of our most potent compounds on group II intron splicing in vivo, we developed a qRT-PCR assay for monitoring the splicing of the ai5γ intron in S. cerevisiae in the presence of small molecules. We found that our most potent compounds cause a severe splicing defect in vivo, which is evident from significant accumulation of precursor RNA molecules containing the 5’-exon-intron junction (Fig. 3b, Supplementary Fig. 4b). Importantly, unspliced COX1 transcripts are targeted for rapid degradation in cells 35, so the direct observation of substantial precursor accumulation suggests a considerable effect on splicing. Taken together, our results in cells demonstrate that the highest-affinity compounds specifically target the ai5γ intron in vivo, selectively disrupting splicing of the COX1 gene and thereby reducing yeast growth.
Selectivity of the group II intron inhibitors.
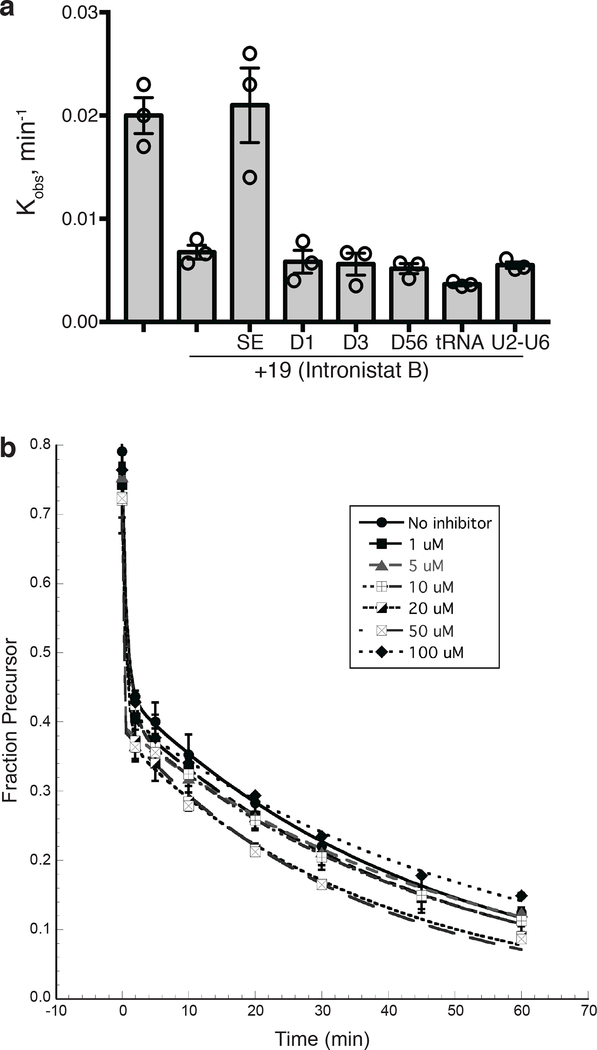
Although the SAR, reversibility analysis and gene specificity of the compounds are consistent with selectivity, it was important to determine whether the compounds can also bind other highly structured RNA molecules, and other RNA splicing systems. In addition, we wondered whether activity requires the fully-folded group II intron RNA tertiary structure, or if individual intron domains can bind the inhibitors with high affinity. To this end, we monitored inhibitory activity of one of the most promising compounds (19) in the presence of a large excess of various RNAs, including separate ai5γ intron domains D1, D3, D56, the U2-U6 snRNA stem-loop (analogous to group II intron D5), and yeast tRNAPhe (Supplementary Fig. 1, see Methods). The latter was an important control because tRNA molecules possess many archetypal elements of RNA tertiary structure, such as kissing loops involving canonical and non-canonical base pairs, coaxially stacked helices, base triples and U-turn motifs, which make them commonly-used specificity controls for RNA targeting 36. However, we observe that none of these RNAs, presented in a 1000-fold excess relative to intron RNA (2 nM), affect the inhibitory activity of compound 19 (Fig. 4a). The only RNA that competed with radiolabeled SE group II intron RNA for binding of 19 was the same unlabeled group IIB intron RNA added in excess (Fig. 4a).
Figure 4.
Active compounds selectively inhibit group II splicing in vitro. (a) Inhibition of the ai5γ group II intron splicing in the presence of excess of different RNAs: unlabeled SE RNA, intronic domains D1, D3 and D56, yeast U2-U6 hairpin and yeast tRNAPhe. The average and s.e.m. (error bars) are calculated from n=3 independent experiments. (b) Group I intron splicing is unaffected by the most potent inhibitor of group IIB intron splicing 19 (Intronistat B). Representative time courses of Azoarcus pre-tRNA (Ile) group I intron splicing in the absence and in the presence of the inhibitor 19 (Intronistat B) at indicated concentrations. Fraction of precursor was quantified at each time point, plotted vs time and fit to a double-exponential equation to determine the reaction rate constants (1 min−1 for the fast population and 0.02 min−1 for the slow population). Data represent average of n=3 independent experiments, error bars are s.e.m.
To evaluate inhibition of the other two known RNA splicing systems (group I and spliceosome) we monitored splicing of the Azoarcus pre-tRNA (Ile) group I intron 37 in the presence of compound 19. We observe that splicing of the Azoarcus intron is unaffected, even at 100 μM compound 19 (Fig. 4b). In addition, we used qRT-PCR to monitor inhibition of group I intron and spliceosomal splicing in S. cerevisiae in vivo. Consistent with our previous results, we found that the inhibitors only affect splicing of the yeast ai5γ group II intron (Fig 3b). Spliceosomal processing, group I intron splicing and even splicing of group II introns from subclasses that differ from subclass IIB are unaffected by the small molecule inhibitors. These results are consistent with the in vitro results and suggest that the inhibitors bind selectively to group IIB introns. Our data also indicate that the inhibitors bind tertiary structural elements formed by the entire intron and not individual intronic domains.
Small molecule growth inhibition of C. parapsilosis.
The yeast pathogen C. parapsilosis contains a single group IIB intron in its COX1 gene 38. The active site of this intron (D5) is almost identical to that of the ai5γ intron (Supplementary Fig. 1b), suggesting that compounds that inhibit the S.c. ai5γ intron may also inhibit splicing by the group II intron in C. parapsilosis. To evaluate efficacy of the compounds against this pathogen, we measured the minimum inhibitory concentrations (MIC values) required for growth inhibition of C. parapsilosis. We observe that the high affinity ai5γ intron inhibitors significantly reduce the growth of C. parapsilosis (Fig. 2, Supplementary table 2). Correlations between IC50, Ki and MIC values suggest that these compounds employ the same mechanism of action both in vitro and in vivo (Fig. 2, Supplementary table 2). While many of the highest-affinity compounds displayed strong MIC values, compounds 18 (Ki = 2.1±0.2 μM) and 19 (Ki = 0.36±0.02 μM) were particularly notable because their MIC values (2–4 μg/ml) are comparable to that of Amphotericin B, which is still commonly used for acute C. parapsilosis infection 39 (MIC is 0.5–1 μg/ml) (Fig. 2, Supplementary table 2). Given their potency and antifungal effects, we have named compound 18 Intronistat A, and compound 19 Intronistat B.
To directly monitor behavior of the compounds in vivo, we used qRT-PCR to quantify levels of splicing for the C. parapsilosis COX1 precursor mRNA (which contains the group IIB intron) in the presence of 19 (Intronistat B) and inactive compound 4. We observed a moderate splicing defect caused by Intronistat B as indicated by increased levels of unspliced C.p.COX1 relative to total, and this effect was not observed in the presence of inactive compound 4 (Supplementary Fig. 4c). To determine whether inhibition is specific to yeast, we evaluated toxicity of the most potent compounds in human cells, determining the IC50 for inhibition of HEK-293T cells (Fig. 2, Supplementary Figure 5, Supplementary table 2). While some of the compounds are broadly toxic to all eukaryotic cells tested, our most potent compounds, including Intronistat A and Intronistat B, did not show toxicity in human cells after 24 h incubation (Fig. 2, Supplementary Fig. 5a, Supplementary table 2). Even after 72h incubation Intronistat B had only mild effects on cell viability (Supplementary Fig. 5b), suggesting that this compound specifically targets yeast strains that contain group II introns in an essential gene and that it lacks cross-reactivity with the nuclear spliceosome, or other targets, in yeast and humans.
Discussion.
In this study, we have demonstrated that the tertiary structure of a large, complex RNA molecule can be targeted de-novo, and that high affinity compounds can be identified using classical methods for developing pharmacologically active compounds. The success of this study implies that high-complexity RNA tertiary structures, such as those found in viral genomes, in the untranslated regions of human mRNAs, pre-miRNA clusters, long intergenic noncoding RNAs, and other RNAs essential for the regulation of gene expression, can potentially be targeted with specific, high-affinity small molecules in order to regulate their function.
An equally significant aspect of this project was the experimental approach utilized for RNA targeting. Most recent examples of RNA targeting by small molecules involve the use of rational drug design, or the serendipitous discovery of natural products 20,40,41. But in this example, we used a classical method that has been refined through decades of investigations aimed at identifying of inhibitors of regulatory proteins such as enzymes and receptor complexes. Indeed, we utilized the primary approach that pharmaceutical companies employ for drug design and optimization. Specifically, we created a high-throughput assay to screen compound libraries for inhibitory activity against an RNA molecule, we identified hits, conducted SAR and then optimized a scaffold to generate potent inhibitors. We then demonstrated target specificity in vitro and in vivo using multiple, orthogonal approaches. The success of this approach is significant for two reasons, particularly given the burgeoning interest in “drugging RNA”. First, it demonstrates that the determinants for RNA recognition are sufficiently similar to those of proteins that existing libraries and strategies can be employed for the identification of high affinity RNA ligands. Second, it demonstrates that de-novo targeting of large complex RNA molecules can readily be accomplished using methods that are well established, and that “reinventing the wheel” will not necessarily be required for identifying inhibitors to bioactive RNA molecules. This is important not least because it makes RNA targeting accessible to a large population of seasoned medicinal chemistry investigators, who have much to contribute in this area of research.
The most potent inhibitors that we identified contain a gallate moiety, which is notable because related polyphenolic compounds, such as catechols, are sometimes found in promiscuous PAINS compounds 42,43. While the presence of a gallate or catechol moiety necessitates caution, it should not disqualify a compound scaffold because catechol reactivity is highly dependent on architecture of the surrounding molecule44. Selectivity and appropriate behavior of such molecules must be established through rigorous SAR, reversibility, binding selectivity, cytotoxicity and other applicable methods, as in this study. Although caution is appropriate, recent studies demonstrate that more than 90% of small molecules that are flagged in automated PAINS screens (such as FAF-drugs3) are well-behaved compounds that have a low hit rate in conventional alpha-screen assays 44,45. Indeed, there is considerable recent concern that valuable bioactive compounds are being removed from the pool of informative chemical space by PAINS screens. This is a particularly serious problem in the development of RNA ligands, as the chemical space for RNA recognition is not well understood, and what is known from studies of RNA binding to macromolecules suggests that base-stacking and H-bonding, particularly to hydroxyls, drives tight binding17,46. For these reasons, the gallate moiety in our group II intron inhibitors may be representative of a useful RNA binding motif, but its inclusion was necessarily accompanied by extensive selectivity analysis.
Finally, this study is significant because of its practical applications. Having shown that group II introns can be targeted with small molecules, we used the resulting compounds to inhibit the growth of yeast pathogens that uniquely depend on group II introns for metabolic function. Indeed, the best inhibitors we identified in this study have MIC values (~2 μg/ml) that are comparable to that of Amphotericin B (0.5–1 μg/ml), which is a drug that is still used to treat severe fungal infections despite serious side effects 39. Consistent with the lack of group II introns in vertebrate animals, our most potent inhibitor Intronistat B is not toxic in mammalian cells and does not influence spliceosomal RNA processing, suggesting that this molecule has potential for further development as an antifungal therapeutic. MIC values for C. parapsilosis are in good agreement with in vitro Ki values for the ai5γ S. cerevisiae intron, indicating that the inhibitors function similarly in different yeast species that contain group II introns, suggesting a route for broad-spectrum fungal inhibitors. Given the unique RNA metabolism of plants, fungi and yeast, our results demonstrate that RNA targeting may provide a much-needed approach for developing therapeutics against eukaryotic pathogens.
Given the vast number of physiologically important RNA tertiary structures that control gene expression in all domains of life, the ability to regulate RNA function with small molecules represents a new frontier in molecular medicine. Here we present a discovery pipeline for targeting RNA tertiary structures de-novo and we demonstrate the pharmacological utility of small molecule modulators by using them to specifically inhibit self-splicing group II introns, thereby disrupting the growth of pathogenic yeast. This demonstrates that RNA tertiary structures are attractive, accessible targets for the development of new probes and therapeutics.
Methods.
Online methods.
Yeast strains.
Strains of Candida parapsilosis (ATCC 22019) and Sacharomyces cerevisiae (ATCC 18824) were purchased from American Type Culture Collection (ATCC) and cultured according to the instructions from the manufacturer (ATCC). S. cerevisiae Wild-type (NP40–36a) and mtDNA intronless (XPM46) strains were kindly provided by Dr. Thomas Fox34.
RNA preparation.
Synthesis of the RNA oligo substrates.
RNA oligonucleotides containing 3’-terminal Black Hole Quencher 2 label and aminomodifier C6dT nucleotide (Glen Research) as well as U2-U6 RNA oligonucleotide (5’ AGC AGU UCC CCU GCAUA AGG AUG AAC CGC U) were synthesized on a MerMade 12 DNA-RNA synthesizer (BioAutomation) using TBDMS phosphoramidites (Glen Research). Base deprotection was carried out in a 3:1 mixture of 30% ammonium hydroxide (JT Baker) and ethanol at room temperature for 24 h. Subsequent 2’-OH deprotection and purification on a 20% denaturing polyacrylamide gel were carried out as previously described 47,48 RNA oligonucleotide U2-U6 was deprotected and purified as described 47,48.
In vitro transcription.
Large-scale transcription of the SE, D1(Domain 1 in isolation), D3 (Domain 3 in isolation), D56 (RNA molecule containing both domains 5 and 6) and D135 RNAs 33,49,50was carried out using T7 RNA polymerase as previously described 51. Internally 32P-labeled ai5γ intron with short exons (SE RNA) 52 was prepared by in vitro transcription as described 53. Internally labeled Azo-Pre-tRNA 37 was in vitro transcribed and purified as previously described 37. RNA molecules D3 and D56 were purified on an 8% denaturing polyacrylamide gel, and all other RNAs were purified on a 5% gel as previously described 48,53.
Fluorescent labeling of RNA oligonucleotides
Purified RNA oligonucleotides containing 3’-terminal Black Hole Quencher 2 label and aminomodifier C6dT nucleotide (Glen Research) were fluorescently labeled with the NHS ester of AlexaFluor 555 dye (Life Technologies Corp.) at the primary amino group located on the aminomodifier C6dT nucleotide. RNA oligonucleotides were dissolved in 200 μl of 0.25 M sodium bicarbonate buffer (pH 9.2) and then combined with a solution containing 0.5 mg of AlexaFluor 555 NHS ester in 200 μl formamide. The reaction was incubated at room temperature for 2 hours and the labelled products were purified on a 20% denaturing polyacrylamide gel.
High-throughput screening.
High-throughput screening was carried out using a library of 10,000 compounds selected from the following collections: NCI Oncology (85 compounds), NCI Diversity (1356 compounds), ENZO kinase inhibitors (80 compounds), 640 FDA approved drugs, ENZO phosphatase inhibitors (33 compounds), BML-ENZO ion channel ligands (72 compounds), BML-metabotropic glutamatergic ligands (56 compounds), BML nuclear receptor ligands (76 compounds), protease inhibitors (53 compounds) and additional compounds from ChemBridge MW and ChemDiv to bring the total number of compounds to 10000.
The D135 ribozyme 33, fluorescently labeled oligo substrate 17/2 DL (Fig. S2) (both at 20 nM final concentrations in 50 mM MOPS, pH 7.0, 100 mM MgCl2, 0.5 M KCl), and the test compounds (final concentration of 10 μM in 50 mM MOPS, pH 7.0, 100 mM MgCl2, 0.5 M KCl) were combined in black non-binding 384-well plates (Corning 3757) using multidrop combi (ThermoFisherScientific) and pintool (V & P Scientific) pipettors. The reaction mixtures were incubated at 37°C for 45 min, and then quenched with 100 mM EDTA. Fluorescence intensity was analyzed on a Tecan Infinite multimode plate reader (λex 520 nm, λem 560 nm, 5nm bandwidth). Data were normalized relative to the untreated wells and to the wells lacking the D135 ribozyme, and percent of inhibition was calculated using ActivityBase (IDBS). The Z’-factor was calculated as described 54
Z’ = 1 – 3 × (σp + σn)/|μp – μn|, where μp and σn are mean value and standard deviation for the positive control (substrate only), and where μn and σn are the mean value and standard deviation for the negative control (no compound). The average Z’-factor from the screen was 0.83±0.03.
IC50 measurement for the small molecule inhibition of the D135 ribozyme cleavage reaction.
Black 96-well plates (Corning 3695) were filled with 50 μl of solution containing 20 nM D135 ribozyme, 20 nM double-labeled substrate 17/2 DL and small molecule inhibitor in 50 mM MOPS, pH 7.0, 100 mM MgCl2 and 500 mM KCl. Small molecule inhibitors were tested at 19 different concentrations ranging from 5 nM to 1 mM. Plates were incubated at 37°C for 10 min., reaction mixtures were quenched with 100 mM EDTA and then analyzed on a Synergy H1 plate reader (BioTek). Each experiment was performed in triplicate. Data were fit to a 4-parameter logistic function c + (d – c)/(1 + (x/a)b), where a is the IC50, b is the slope parameter, c is the minimum response and d is the maximum response. Data are reported as average ± s.e.m
Determination of Ki values for small molecule inhibition of the self-splicing reaction.
Internally labeled SE RNA (2 nM) was incubated with various concentrations of inhibitor compound under near-physiological conditions (in 50 mM MOPS, pH 7.5, 8 mM MgCl2, 100 mM KCl) at 30°C. Aliquots at different time points were quenched and analyzed on a 5% denaturing polyacrylamide gel as previously described 53. Data were fit to a single-exponential equation to determine the first order rate constants (kobs). The latter were plotted against the concentration of inhibitor and fit to the equation for non-competitive inhibition to determine Ki values:
kobs = kmax/(1+[I]/Ki), where kobs and kmax are the first order rate constants measured in the presence and in the absence of the inhibitor, respectively, [I] is the concentration of the inhibitor and Ki is the inhibition constant. Experiments were performed four times for compound 3, three times for 14, 18 (Intronistat A), and 19 (Intronistat B) and twice for the remaining compounds to ensure reproducibility. Data represent average ± s.e.m.
Effect of excess of various RNAs on splicing inhibition.
Internally labeled SE RNA (2 nM final concentration) and unlabeled SE, D1, D3, D56 RNAs, yeast tRNA Phe and U2-U6 RNA oligo (2 μM final concentrations) were preincubated separately in 50 mM MOPS, pH 7.5, 100 mM KCl and 5 mM MgCl2 (10 mM MgCl2 for the tRNA) at 30°C for 20 min. Then the labeled SE RNA and competitor RNA solutions were mixed together with simultaneous addition of 19 (Intronistat B) (300 nM final concentration) to initiate the reaction. Reaction was carried out under near-physiological conditions (in 50 mM MOPS, pH 7.5, 8 mM MgCl2, 100 mM KCl) at 30°C. Aliquots at different time points were quenched and analyzed on an 5% denaturing polyacrylamide gel as previously described 53. Data were fit with a single-exponential equation to determine the first order rate constants (kobs). Experiments were performed in triplicate to ensure reproducibility.
Testing reversibility of compound binding.
To examine reversibility of compound binding, the splicing reaction was initiated at high concentration (2 μM) of the indicated inhibitor, allowed to react for an hour under near-physiological conditions (see above), and then diluted by 10-fold with reaction buffer. Aliquots at different time points were quenched and analyzed on an 5% denaturing polyacrylamide gel as previously described 53. Splicing efficiency was observed to significantly increase upon dilution of the inhibitor, with a change in rate that is commensurate with the reduction in compound concentration, indicating that binding of the inhibitors to the intron RNA is reversible (Supplementary Fig. 3d). Experiments were performed in duplicate to ensure reproducibility.
Inhibition of group I intron splicing.
Splicing of the Azo-Pre-tRNA intron was carried out essentially as previously described 37. Internally labeled Azo-Pre-tRNA (20 nM) was incubated in 25 mM HEPES, pH 7.5, 10 mM MgCl2 at 50 °C for 10 min, and then at 32 °C for 2 min. Then 19 (Intronistat B) was added to the final concentration of 1, 5, 10, 20, 50 and 100 μM and reaction was initiated by addition of 100 μM GTP (final concentration). The final concentration of DMSO in all samples was 10%. Aliquots at different time points were quenched and analyzed on an 5% denaturing polyacrylamide gel as previously described 37,53. All experiments were performed in triplicate to ensure reproducibility. Data represent average ± s.e.m.
Yeast respiration assay in S. cerevisiae.
Yeast respiration assays were conducted either in liquid YPD media (BD Bacto Yeast Extract, BD Bacto Peptone, 2% glucose) or in YPGE media (BD Bacto Yeast Extract, BD Bacto Peptone, 3% glycerol, 3% ethanol), essentially as described for the antifungal MIC assays (see below) using the guidelines from the Clinical and Laboratory Standards Institute 55. To initiate these experiments, 100 μl of a fresh stock solution was prepared for each compound (3.2 mg/ml for Amphotericin B and 12.8 mg/ml for each test compound) in DMSO. This concentrated stock solution was placed in the first well of a 12-well row on a 96-well plate. The subsequent nine wells in this row contained 50 μl of DMSO for serial dilution. From the first well, 50 μl of the compound stock was withdrawn, transferred to the second well and mixed. This process was repeated from the second and subsequent wells, resulting in a 1:2 serial dilution of the stock into DMSO. Wells 11 and 12 contained DMSO only, to be used as the no-compound control and sterility control, with no inoculum added, respectively. After preparing this plate, 2.5 μl from each of the 12 wells was transferred to the wells of a separate 96-well plate, each of which contained 122.5 μl of YPD or YPGE medium, resulting in a set of serial dilutions of compound into growth media in a volume of 125 μl. Then 50 μl from each of these dilutions were transferred onto a third 96-well plate (the assay plate) and mixed with an equal volume (50 μl) of freshly prepared yeast inoculum in YPD or YPGE medium set to provide 0.5×103 to 2.5×103 colony forming units per 1 mL (wells 1–11), or YPD/YPGE alone (sterility control, well 12). The final compound concentration range of the assay was 0.25 – 128 μg/ml for test compounds and 0.0625 – 32 μg/ml for Amphotericin B and 1% DMSO. Plates were incubated at 30°C and visually analyzed for turbidity after 24 and 48 h of incubation. The lowest compound concentration at which no growth was apparent is reported as the MIC (Supplementary Table 3). All experiments were performed in triplicate.
S. cerevisiae growth assays
Cultures of S. cerevisiae (Wild-type: NP40–36a, mtDNA intronless: XPM46 34) were grown overnight with shaking to mid-log phase in YPGE (see above) at 30°C. Equal numbers of cells were harvested and serially-diluted 1:5. Cells were then plated on YPD or YPGE media containing DMSO or 18 (Intronistat A) dissolved in DMSO (final concentration: 64 or 128μg/mL). Plates were grown for one day (YPD plates), two days (YPGE DMSO plate), or 18 days (YPGE 18 (Intronistat A) plate) at 30°C. Experiment was replicated twice to ensure reproducibility of growth phenotype.
Analysis of splicing in S. cerevisiae by qRT-PCR.
A culture of S. cerevisiae was grown overnight with shaking in YPGE (see above) at 30 °C. The saturated culture was then diluted and grown to mid-log phase in YPGE. Equal volumes of DMSO or compounds dissolved in DMSO were added to individual cultures (final concentrations of 19 (Intronistat B): 64μg/mL, 18 (Intronistat A): 32μg/mL, 12: 32 μg/mL, 3: 64 μg/mL, Amphotericin B: 8 μg/mL), and incubated for four hours. Cells were harvested by centrifugation, washed with 500 uL ice-cold MilliQ water, centrifuged again, and snap-frozen in liquid nitrogen. Total RNA was isolated using E.Z.N.A. Yeast Purification Kit (Omega Bio-tek) according to manufacturer’s procedures. The RNA was eluted in 50 uL of RNase/DNase-free water (ThermoFisher), followed by DNAse treatment with RQ1 DNase (Promega), for 1hr at 37°C. Then it was mixed with 6 μl of 3M NaOAc and precipitated with 75% EtOH at −20°C overnight. Then the RNA was reverse-transcribed with SuperScript III (ThermoFisher) following manufacturer’s recommendations with 200 ng Random Hexamer Primers (ThermoFisher) and 40U RNasin (ThermoFisher) in a 20 μl reaction. After reverse transcription, RNA was degraded by addition of 2N NaOH (2 μl ) and incubation at 95 °C for 5 min. After cooling on ice for 5 min, 2 μl of 1M HCl and 2 μl of 3M NaOAc were added to the reaction mixture, and cDNA was precipitated with 75% EtOH at −20°C for 30 min and resuspended in RNase/DNase-free water. The cDNA levels were quantified with real-time PCR using LightCycler 480 SYBR Green I (Roche) and a CFX384 Real-Time PCR Detection System (Bio-Rad) in triplicate with independent samples. Relative levels of indicated RNA species from different conditions were normalized according to the ΔΔCT method via the following equation 56
ΔΔCT = [(CT COX1 total or unspliced -CT ACT1 or PGK1) test compound - (CT COX1 total or unspliced - CT ACT1 or PGK1) DMSO)]
The mean and s.e.m. of 2^-ΔΔCT is reported for each sample from four independent replicates. The following primer sets were used to amplify indicated targets: ACT1: TCGAACAAGAAATGCAAACCG, GGCAGATTCCAAACCCAAAAC; PGK1: TGTCTTGGCTTCTCACTTGG, TTCAACTTCTGGACCGACAC; total COX1: TGGTATGCCTAGAAGAATTCCTG, AGAATAATGATAATAGTGCAATGAATGAAC; COX1 unspliced aI1: GGCAGGAACAGCAATGTCTT, TGCTAGACGCATCAACGAAA; COX1 unspliced aI2: TTAGTAGTTGGTCATGCTGTATT, CCCAACTGGGTAAGCCATA; COX1 unspliced aI5α: TGCTATGGCTTCAATTGGATT, TTTTATTTTTATTTTTATCCTTGCTAAAGGG; COX1 unspliced aI5β: TCACAATGGGTGGTTTAACTGG, ATTTTAGTGAATTTTAAGCACGACAC; COX1 unspliced aI5γ: CTTACTACGTGGTGGGACATT, GTCATTACAGCTTAGCATATTTATGT; total YRA1: AAACGCAGTCGCTAGAGTTG, CCTGCTTAATGTCCCTTGGC; unspliced YRA1: GTCAAGGTCAACGTCGAAGG, CACGAGACGATGCGAGTAAC; total MTR2: ACACAACATGCCCTAACAGG, TTGGGTCGATTTCTGTTCCC; unspliced MTR2: GGATGCGCAAACGCATAG, TCTTCGTAAATGTGGCCGTT. In the course of analysis, no RT control samples displayed minimal signal, indicating that amplicons had been generated from cDNA and not genomic DNA, and gel electrophoresis confirmed specific amplification of the intended amplicon.
Analysis of group II intron splicing in C. parapsilosis by qRT-PCR.
A culture of C. parapsilosis was grown overnight with shaking in RPMI at 30 °C. The saturated culture was then diluted and grown to mid-log phase in RPMI. Equal volumes of DMSO or compounds dissolved in DMSO were added to individual cultures (final concentrations of Intronistat B (19) and 4: 32μg/mL) and incubated for two hours. Potassium cyanide (KCN), an inhibitor of complex IV of the electron transport chain, was added to a final concentration of 10 mM for an additional two hours to induce nascent COX1 expression. Cells were harvested, RNA prepared and reverse-transcribed, and cDNA quantified as in S. cerevisiae experiments. The mean and s.e.m. of 2^-ΔΔCT is reported for each sample from three independent replicates. The following primer sets were used to amplify indicated targets: C.p.PGK1: TGGATGGGTCTTGATTGTGG, GTCAAATTCAAAGACACCCGG; total C.p.COX1: GGTGCTGTAGATATGGCATTTG, GCACTAATTGATGATAGTGGAGGA; unspliced C.p.COX1: TGTTCTTGTTACTGGTCATGCT, AGCACTTACTAACTGTTCACGTC.
Determination of MICs for the small molecule inhibitors against C. parapsilosis
Minimal inhibitory concentrations (MICs) for C. parapsilosis were determined according to the guidelines from the protocol M27-A3 from the Clinical and Laboratory Standards Institute (CLSI) 55 in a final volume of 100 μl. Fresh stock solutions of control antifungal drugs (Amphotericin B and Itraconazole) and of test compounds were prepared at 3.2 mg/ml and at 12.8 mg/ml respectively in DMSO. Compound dilution plates and assay plates were prepared as described above for the yeast respiration assay, except that RPMI-1640 medium (Sigma) was used instead of YPD or YPGE media. The use of RPMI-1640 medium in antifungal MIC experiments is mandated by the CLSI protocol. Unlike S. cerevisiae, which only respires in glycerol and ethanol media, C. parapsilosis respires in both glycerol and glucose media and demonstrates severe growth defects when respiration is limited 31. Therefore a C. parapsilosis growth defect in RPMI-1640 medium would be expected in the presence of the group II intron splicing inhibitors.
The final concentration range of the assay was 0.25 – 128 μg/ml for test compounds and 0.0625 – 32 μg/ml for Amphotericin B and Itraconazole. The final concentration of DMSO was 1%. The assay plate was incubated at 35°C or 37°C and visually analyzed for turbidity after 24 and 48 h of incubation. The lowest compound concentration at which no growth was apparent is reported as the MIC. All experiments were performed in triplicate.
Cytotoxicity in HEK-293T cells.
HEK-293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco) supplemented with 10% Fetal Bovine Serum (FBS, Gibco) and 100 U/ml of Penicillin-Streptomycin (Gibco) at 37 °C and 5% CO2. For the cytotoxicity experiments, cells were aliquoted into black 96-well plates with a clear bottom (Corning 3603) at a concentration of 10,000 cells per well. The cells were grown at 37°C, 5% CO2 for 5–6 hours, then the medium was replaced with the same medium without FBS and cells were grown at 37°C, 5% CO2 for 24 hours. Freshly prepared stock solutions of test compounds (12.8 mg/ml in DMSO) were serially diluted 1:2 with DMSO into the first 11 successive wells of a 12-well row on a 96-well plate as described above. Well 12 was used as a growth control (no compound). After dilution, 1 μl from each well of the compound plate was added to the plate containing cells (the assay plate), and cells were incubated at 37°C, 5% CO2 for another 24 or 72 hours. After incubation, cell viability was determined using the luminescent Cell Titer Glo cell viability assay (Promega), in which 100 μl of the assay reagent was added to each well of the assay plate. After gentle shaking for 7–10 minutes, the plates were analyzed on a Synergy H1 plate reader (BioTek). Luminescence was plotted against the compound concentration and IC50 values were determined by fitting the data to a 4-parameter logistic function c + (d – c)/(1 + (x/a)b), where a is the IC50, b is the slope parameter, c is the minimum response and d is the maximum response. Experiments were replicated 4 times for compounds 2, 3, 5, 7, 9, 11, 12, 15, Intronistat A (18) and Intronistat B (19), 3 times for compounds 1, 6, 8, 10, 13, 14, 16 and 17, and twice for compound 4 to ensure reproducibility.
Statistics and reproducibility.
The following reported data represent average of n=3 independent experiments: all IC50 values for ribozyme cleavage, Ki values for compounds 14, 18 (Intronistat A) and 19 (Intronstat B), kobs values for inhibition of splicing in the presence of the excess of various RNAs, time courses for inhibition of the group I intron splicing, ΔΔCT values obtained from qRT-PCR analysis of group II intron splicing in C. parapsilosis, MIC values from yeast respiration assay, MIC values for small molecules against C. parapsilosis, IC50 values for cytotoxicity in HEK-293T cells for compounds 1, 6, 8, 10, 13, 14, 16 and 17.
The following reported data represent average of n=4 independent experiments: Ki for compound 3, ΔΔCT values obtained from qRT-PCR analysis of group II intron splicing in S. cerevisiae, IC50 values for cytotoxicity in HEK-293T cells for compounds 2, 3, 5, 7, 9, 11, 12, 15, Intronistat A (18) and Intronstat B (19).
The following experiments were repeated twice to ensure reproducibility: test of reversibility of compound binding, S. cerevisiae growth assays, cytotoxicity of compound 4 in HEK-293T cells, Ki determination for compounds 1, 2, 4–13 and 15–17.
All values are reported as mean ± s.e.m.
Supplementary Material
Acknowledgements.
We thank Sheila Umlauf, Dr. Peter Gareiss and Dr. Jane Merkel at Yale Center for Molecular Discovery for their help with high throughput screening. We are grateful to Dr. Kenneth Blount and Dr. Ronald Breaker for help with setting up MIC expriments. We thank Dr. Julie Sinclair and Dr. Alanna Schepartz for sharing their expertise on cytotoxicity experiments. We gratefully acknowledge Dr. Thomas Fox for sharing wild-type and mtDNA intronless S. cerevisiae strains. We thank Dr. Sarah Woodson for sharing the Azo-pre-tRNA plasmid. We thank Dr. David Chenoweth and Dr. Albert DeBerardinis for helpful discussions. We are grateful to Dr. Seth Herzon and Dr. Ronald Breaker for comments on the manuscript. We are grateful to Dr. Chen Zhao for help in making Supplementary Fig. 1. A.M.P. is an Investigator and O.F is a Research Specialist in the Howard Hughes Medical Institute. This work was supported by NIH grant RO1GM50313 to A.M.P. and R43 AI115951 to M.V.Z.
Footnotes
Competing financial interest statement.
Yale University has filed a provisional patent application on the work developed in this manuscript.
Data availability statement.
Authors can confirm that all relevant data are included in the paper and/or its supplementary information files.
References
- 1.Blount KF & Breaker RR Riboswitches as antibacterial drug targets. Nat Biotechnol 24, 1558–64 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Howe JA et al. Selective small-molecule inhibition of an RNA structural element. Nature 526, 672–7 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Howe JA et al. Atomic resolution mechanistic studies of ribocil: A highly selective unnatural ligand mimic of the E. coli FMN riboswitch. RNA Biol 13, 946–954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H et al. Dual-Targeting Small-Molecule Inhibitors of the Staphylococcus aureus FMN Riboswitch Disrupt Riboflavin Homeostasis in an Infectious Setting. Cell Chem Biol 24, 576–588 e6 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Kim JN et al. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem Biol 4, 915–27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blount KF, Wang JX, Lim J, Sudarsan N & Breaker RR Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol 3, 44–9 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Blount KF et al. Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob Agents Chemother 59, 5736–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Ahsen U, Davies J & Schroeder R Antibiotic inhibition of group I ribozyme function. Nature 353, 368–70 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Disney MD, Childs JL & Turner DH Hoechst 33258 selectively inhibits group I intron self-splicing by affecting RNA folding. Chembiochem 5, 1647–52 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Mei HY, Cui M, Lemrow SM & Czarnik AW Discovery of selective, small-molecule inhibitors of RNA complexes--II. Self-splicing group I intron ribozyme. Bioorg Med Chem 5, 1185–95 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Patwardhan NN et al. Amiloride as a new RNA-binding scaffold with activity against HIV-1 TAR. Medchemcomm 8, 1022–1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder R, Waldsich C & Wank H Modulation of RNA function by aminoglycoside antibiotics. EMBO J 19, 1–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colameco S & Elliot MA Non-coding RNAs as antibiotic targets. Biochem Pharmacol 133, 29–42 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Wilson DN Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12, 35–48 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Blaha GM, Polikanov YS & Steitz TA Elements of ribosomal drug resistance and specificity. Curr Opin Struct Biol 22, 750–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eubanks CS, Forte JE, Kapral GJ & Hargrove AE Small Molecule-Based Pattern Recognition To Classify RNA Structure. J Am Chem Soc 139, 409–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan BS, Forte JE, Culver RN, Zhang Y & Hargrove AE Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA-Targeted Bioactive Ligands. Angew Chem Int Ed Engl 56, 13498–13502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernat V & Disney MD RNA Structures as Mediators of Neurological Diseases and as Drug Targets. Neuron 87, 28–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costales MG et al. Small Molecule Inhibition of microRNA-210 Reprograms an Oncogenic Hypoxic Circuit. J Am Chem Soc 139, 3446–3455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang WY, Gao R, Southern M, Sarkar PS & Disney MD Design of a bioactive small molecule that targets r(AUUCU) repeats in spinocerebellar ataxia 10. Nat Commun 7, 11647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dibrov SM et al. Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J Med Chem 57, 1694–707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barros SA, Yoon I & Chenoweth DM Modulation of the E. coli rpoH Temperature Sensor with Triptycene-Based Small Molecules. Angew Chem Int Ed Engl 55, 8258–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sztuba-Solinska J et al. Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J Am Chem Soc 136, 8402–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz DA, Song JM & Garner AL High-throughput platform assay technology for the discovery of pre-microrna-selective small molecule probes. Bioconjug Chem 26, 19–23 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Kett DH, Azoulay E, Echeverria PM, Vincent JL & Extended Prevalence of Infection in, I.C.U.S.G.o.I. Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Care Med 39, 665–70 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Guinea J Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20 Suppl 6, 5–10 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Morales DK et al. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 4, e00526–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcia M & Pyle AM Principles of ion recognition in RNA: insights from the group II intron structures. RNA 20, 516–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao C & Pyle AM Structural Insights into the Mechanism of Group II Intron Splicing. Trends Biochem Sci 42, 470–482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perlman PS Genetic analysis of RNA splicing in yeast mitochondria. Methods Enzymol 181, 539–58 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Rossignol T et al. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot Cell 8, 550–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard ML, Nobile CJ, Bruno VM & Mitchell AP Candida albicans biofilm-defective mutants. Eukaryot Cell 4, 1493–502 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su LJ, Waldsich C & Pyle AM An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res 33, 6674–87 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Martinez X, Broadley SA & Fox TD Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J 22, 5951–61 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dziembowski A et al. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem 278, 1603–11 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Luedtke NW, Liu Q & Tor Y RNA-ligand interactions: affinity and specificity of aminoglycoside dimers and acridine conjugates to the HIV-1 Rev response element. Biochemistry 42, 11391–403 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Tanner M & Cech T Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA 2, 74–83 (1996). [PMC free article] [PubMed] [Google Scholar]
- 38.Li CF, Costa M, Bassi G, Lai YK & Michel F Recurrent insertion of 5’-terminal nucleotides and loss of the branchpoint motif in lineages of group II introns inserted in mitochondrial preribosomal RNAs. RNA 17, 1321–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moen MD, Lyseng-Williamson KA & Scott LJ Liposomal amphotericin B: a review of its use as empirical therapy in febrile neutropenia and in the treatment of invasive fungal infections. Drugs 69, 361–92 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Velagapudi SP et al. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci U S A 113, 5898–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulhbacher J et al. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog 6, e1000865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baell J & Walters MA Chemistry: Chemical con artists foil drug discovery. Nature 513, 481–3 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Baell JB & Nissink JWM Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017-Utility and Limitations. ACS Chem Biol 13, 36–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jasial S, Hu Y & Bajorath J How Frequently Are Pan-Assay Interference Compounds Active? Large-Scale Analysis of Screening Data Reveals Diverse Activity Profiles, Low Global Hit Frequency, and Many Consistently Inactive Compounds. J Med Chem 60, 3879–3886 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Capuzzi SJ, Muratov EN & Tropsha A Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS. J Chem Inf Model 57, 417–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Re A, Joshi T, Kulberkyte E, Morris Q & Workman CT RNA-protein interactions: an overview. Methods Mol Biol 1097, 491–521 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Wincott F et al. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res 23, 2677–84 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickey TH & Pyle AM The SMAD3 transcription factor binds complex RNA structures with high affinity. Nucleic Acids Res 45, 11980–11988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin K & Pyle AM Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5’-splice site selection. RNA 1, 391–406 (1995). [PMC free article] [PubMed] [Google Scholar]
- 50.Fedorova O, Mitros T & Pyle AM Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol 330, 197–209 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Pyle AM & Green JB Building a kinetic framework for group II intron ribozyme activity: quantitation of interdomain binding and reaction rate. Biochemistry 33, 2716–25 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Zingler N, Solem A & Pyle AM Dual roles for the Mss116 cofactor during splicing of the ai5gamma group II intron. Nucleic Acids Res 38, 6602–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daniels DL, Michels WJ Jr. & Pyle AM Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J Mol Biol 256, 31–49 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Zhang JH, Chung TD & Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 55.CLSI. Reference Method For Broth Dilution Antifungal Susceptibility Testing of yeasts. Approved standard M27-A3, (Wayne, PA, 2008).
- 56.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.