Abstract
DNA methylation patterns regulate gene expression programs and are maintained through a highly coordinated process orchestrated by the RING E3 ubiquitin ligase UHRF1. UHRF1 controls DNA methylation inheritance by reading epigenetic modifications to histones and DNA to activate histone H3 ubiquitylation. Here, we find that all five domains of UHRF1, including the previously uncharacterized ubiquitin-like domain (UBL), cooperate for hemi-methylated DNA-dependent H3 ubiquitin ligation. Our structural and biochemical studies, including mutations found in cancer genomes, reveal a bifunctional requirement for the UBL in histone modification: (1) the UBL makes an essential interaction with the backside of the E2 and (2) the UBL coordinates with other UHRF1 domains that recognize epigenetic marks on DNA and histone H3 to direct ubiquitin to H3. Finally, we show UBLs from other E3s also have a conserved interaction with the E2, Ube2D, highlighting a potential prevalence of interactions between UBLs and E2s.
Keywords: UHRF1, ubiquitin, Ubiquitin-like domain, UBL, DNA methylation, epigenetics, histone H3 ubiquitylation, hemi-methylated DNA, RING E3 ligase, SRA, TTD, PHD, RING
Graphical Abstract
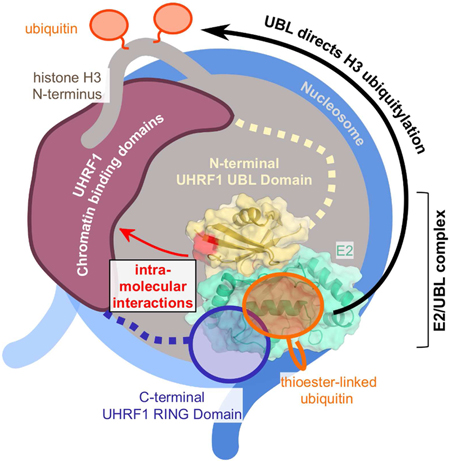
INTRODUCTION:
DNA methylation patterns are established early in development and are an important transcriptional regulatory signal (Smith and Meissner, 2013). In mammals, methylation most commonly occurs symmetrically on cytosines at CpG dinucleotides, marking 70–80% of CpG sites in the genome (Ehrlich et al., 1982). When cells divide, the DNA methylation patterns must be faithfully passed to daughter cells to prevent aberrant changes in gene expression and to retain cell-type identity. During DNA replication, the daughter strand is synthesized without cytosine methylation (Figure 1A, left). Replication-coupled inheritance of methyl-CpG dinucleotides requires the DNA methyltransferase, DNMT1, which methylates the unmodified cytosine bases on the daughter strands to preserve the epigenetic mark (Figure 1A, right) (Goyal et al., 2006; Hervouet et al., 2018). However, DNMT1 cannot perform its function without the RING E3 ligase UHRF1 (Bostick et al., 2007; Rothbart et al., 2012a; Sharif et al., 2007). UHRF1 responds directly to asymmetrically methylated DNA (hemi-methylated; he5mc) and dictates DNA methylation inheritance through histone H3 ubiquitylation (Figure 1A, middle) (Harrison et al., 2016a). UHRF1-generated ubiquitylated H3 (H3ub) acts as a recruitment and activation signal for the DNA methyltransferase, DNMT1 (Figure 1A, right) (Ishiyama et al., 2017; Nishiyama et al., 2013; Qin et al., 2015). Little is known about the mechanism by which this remarkable RING E3 ligase translates a he5mc signal into a critical epigenetic mark.
Figure 1 |. UHRF1 is a multi-domain protein that ubiquitylates histone H3.
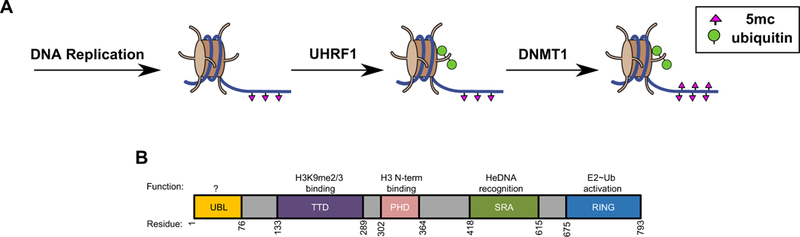
(A) Following DNA replication, UHRF1 recognizes hemi-methylated DNA sites and attaches mono-ubiquitin to Lys 14, 18, 23, and 27 on the N-terminal tail of histone H3 (Harrison et al., 2016a), to serve as a recruitment signal for DNA methyltransferase DNMT1.
(B) Domain architecture of UHRF1 and approximate domain boundaries: UBL, ubiquitin-like domain; TTD, tandem tudor domain; PHD, plant homeodomain; SRA, SET and RING finger-associated domain; RING, really interesting new gene. Previously determined functions are displayed above each domain.
While the mechanism of UHRF1 action remains elusive, general RING-type E3 ligase mechanisms have been defined. Typically, RING-type E3s, the largest family of eukaryotic E3, simultaneously bind a substrate and an E2-ubiquitin conjugate (E2~Ub) to effect the direct transfer of ubiquitin (Ub) from the E2 active site to a lysine residue of a substrate protein. A mechanism by which RING domains activate E2~Ub conjugates for Ub transfer has been defined for the Ube2D family of E2s (Supplemental Figure 1A) including the human paralogues Ube2D1, 2, and 3, (also known as UbcH5a, b, and c and collectively referred to here as “Ube2D” (Dou et al., 2012; Plechanovová et al., 2012; Pruneda et al., 2012)).
While RING domains have the key function of binding and activating E2~Ub conjugates, we have previously shown that UHRF1 requires cooperation among its functional domains to coordinate Ub ligation to H3. Functions of four of the five UHRF1 domains have been defined (Figure 1B): the epigenetic status of histone H3 is sensed by the linked Tandem Tudor Domain (TTD) and Plant Homeodomain (PHD), which together bind tightly to H3 N-termini with a preference for those carrying a K9me2/3 modification (Rajakumara et al., 2011; Rothbart et al., 2013). The methylation status of DNA is read by the SET and RING-associated domain (SRA), a DNA-binding domain that has higher binding affinity for DNA containing a he5mc (Arita et al., 2008; Avvakumov et al., 2008; Hashimoto et al., 2008). Lastly, the RING domain imparts E3 Ub ligase activity. We recently discovered that UHRF1 binding to he5mc is an allosteric trigger that robustly stimulates the formation of H3ub species (Harrison et al., 2016a) - connecting the histone-binding, DNA-binding, and ubiquitin ligase activity to DNA methylation inheritance.
Missing from this picture is the N-terminal UBL domain. Here we demonstrate a bifunctional requirement for the UHRF1-UBL in the formation of H3ub conjugates. First, the UBL domain binds to the E2 Ube2D with greater affinity than does the UHRF1-RING domain. This finding is unexpected because RING domains are usually the primary E2-recruitment modules within E3s. The UHRF1-UBL binds on the surface opposite the Ube2D active site known as the “backside” that is known to bind to Ub (Brzovic et al., 2006). (Supplemental Figure 1A). Although the UHRF1-UBL and Ub share a β-grasp fold and bind to the same surface of Ube2D, we find that formation of a UHRF1-UBL/Ube2D complex produces distinct effects to those of Ub/Ube2D binding. Second, we have discovered surfaces of the UHRF1-UBL that are not involved in E2 binding but are essential for directing ubiquitin to histone H3 based on mutations in UHRF1-UBL that retain auto-ubiquitylation and E2 binding but are defective in histone H3 ubiquitylation. We define a dual role for the UBL domain of UHRF1 to both recruit E2~Ub to the site of he5mc and direct the specific attachment of Ub to histone H3. We also characterize mutations in the UBL found in cancer genomes further defining a role for UHRF1 in disease.
RESULTS:
The UHRF1-UBL binds to Ube2D and is required for E3 activity
Because no function had yet been assigned to the UHRF1-UBL, we sought to determine its role. Using an in vitro reconstituted ubiquitylation assay containing human UHRF1 and H3-peptide (residues 1–32), we observe robust H3-peptide ubiquitylation only in the presence of DNA oligos that contain a hemi-methylated CpG dinucleotide (he5mc) (Figure 2A, middle lanes). We observe an identical pattern of H3ub products (H3ub1–4) when using a ubiquitin variant that lacks lysines (Ub-K0) and is unable to form poly-ubiquitin chains (Supplemental Figure 1B). These results confirm our previous study showing that UHRF1 adds multiple mono-ubiquitin marks to four sites on the H3-peptide (K14, K18, K23, and K27) (Harrison et al.,2016a). However, N-terminally truncated UHRF1 that lacks the UBL domain (ΔUBL; residues 125–793) displayed no ubiquitylation activity, demonstrating a requirement of the UHRF1-UBL for ligase activity (Figure 2A, right lanes).
Figure 2 |. The UBL domain of UHRF1 binds to the Ube2D family of E2s.
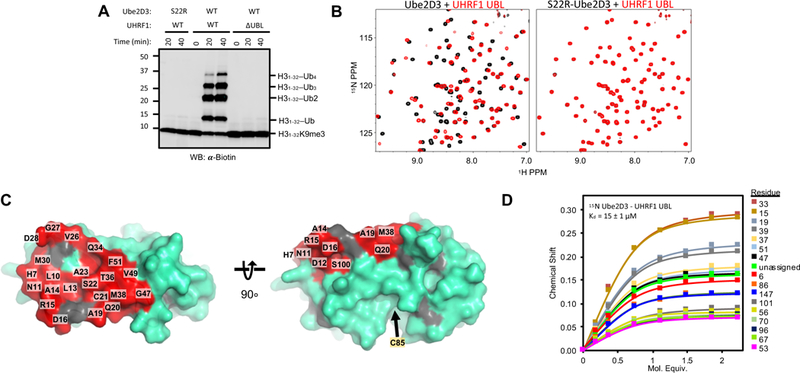
(A) Substrate (H31–32K9me2; H31–32) ubiquitylation assay in the presence of he5mc-DNA with wild-type (WT) or S22R mutant E2 (Ube2D3) and WT UHRF1 or UHRF1 lacking the UBL domain (ΔUBL). Western blot is for the biotin tag on H31–32K9me2.
(B) 1H15N-HSQC spectra of 15N Ube2D3 (C85S; 150 μM) (left) or 15N Ube2D3 (C85S/S22R; 150 μM) (right) in the absence (black) or presence (red) of UHRF1-UBL (150 μM).
(C) Surface representation of the E2 Ube2D (PDB 4V3L) with the UHRF1-UBL-binding surface as determined from NMR shown in red. Highlighted residues correspond to NMR peaks with chemical shift perturbations and intensity reductions (upon complex formation) greater than one standard deviation of the average shift/intensity loss (see Supplemental Figure 1B). Gray, prolines.
(D) Binding curves generated from 1H15N-HSQC peak chemical shifts of 150 μM 15N-Ube2D3(C85S) as a function of [UHRF1-UBL] for individual resonances. See methods for details of Kd determination.
Based on the high sequence homology between Ub and the UHRF1-UBL (Harrison et al., 2015), we suspected the UBL might interact with the E2, Ube2D, in a manner similar to Ub (Brzovic et al., 2006). To test for a direct interaction, we collected 1H15N-HSQC NMR spectra of 15N-Ube2D3 in the presence of increasing unlabeled UHRF1-UBL (residues 1–76). Extensive perturbations were observed in the Ube2D3 NMR spectrum upon addition of the UBL (Figure 2B, left and Supplemental Figure 1C) that define the Ube2D3 surface that binds the UHRF1-UBL, as mapped in red (Figure 2C). The interaction surface corresponds to the backside β-sheet of Ube2D3 and closely overlaps with the surface perturbed upon addition of Ub (Supplemental Figure 1D) (Brzovic et al., 2006; Buetow et al., 2015). A fit of chemical shift perturbations (CSPs) as a function of UHRF1-UBL concentration yields a Kd of 15 ± 1 μM (Figure 2D), substantially tighter than the Kd of ~200–300 μM for the Ub/Ube2D interaction (Brzovic et al., 2006; Buetow et al., 2015).
Mutation of Ube2D residue Ser 22 to arginine (S22R) is known to abrogate Ub binding and NMR experiments confirm that S22R-Ube2D3 no longer binds UHRF1-UBL (Figure 2B, right). Notably, S22R-Ube2D is severely defective in H3 ubiquitylation (Figure 2A, left lanes), indicating that the UBL/E2 interaction plays a central role in UHRF1 ligase activity.
UHRF1 interacts with Ube2D family E2s
We used a previously described yeast two-hybrid assay (Christensen et al., 2007) with thirty human E2s each fused to a transcription-activating domain and the UHRF1-UBL fused to a DNA-binding domain to identify human E2s that bind to UHRF1-UBL. In the assay, growth of histidine auxotrophic yeast is supported if the UBL and an E2 interact. UBL-dependent growth was supported only by Ube2D family members (1, 2, and 3) (Supplemental Figure 2A-C). Similarly, H3-peptide ubiquitylation was observed only with Ube2D family members in assays using full-length UHRF1 with a subset of E2s (Supplemental Figure 2D). There is a modest preference for Ube2D2 and Ube2D3 paralogues in both the yeast two-hybrid and ubiquitylation assays (Supplemental Figure 2).
The UHRF1-UBL and Ub interact with the same surface of Ube2D
To further define the interaction between the UHRF1-UBL and Ube2D we used a combination of NMR spectroscopy, structural modeling, and mutational studies. NMR binding experiments, described above, defined the UBL-binding surface on Ube2D. The UHRF1-UBL spectrum was assigned using standard triple-resonance protocols (see Methods), allowing identification of resonances in the 15N-UHRF1-UBL spectrum perturbed by addition of Ube2D (Figure 3A, right; Supplemental Figure 3A). The perturbed surface is composed of the β-sheet of the UHRF1-UBL and is structurally analogous to the hydrophobic patch centered around isoleucine 44 that Ub uses to interact with the backside of Ube2D (Brzovic et al., 2006) and with most other Ub-binding partners (Harrison et al., 2016b; Winget and Mayor, 2010).
Figure 3 |. Generation of a UHRF1-UBL/Ube2D complex model.
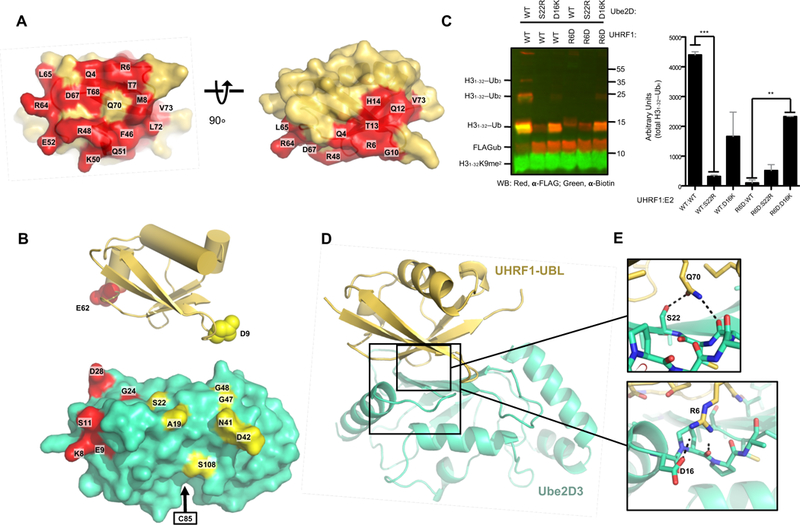
(A) Surface representation of the UHRF1-UBL (PDB 2FAZ) with the NMR-determined E2-binding surface shown in red as per methodology in Figure 2C (see also Supplemental Figure 3A).
(B) UHRF1-UBL structure showing residues modified with the paramagnetic spin label TEMPO (top) and the Ube2D residues affected by the spin labels (bottom) for E62C-TEMPO (red) and D9C-TEMPO (yellow) on the surface of the E2 (PDB 4V3L). See methods and Supplemental Figure 3B and C).
(C) Western blot (left) and quantification (right) of substrate (H31–32K9me2; H31–32) ubiquitylation reactions between indicated pairs of Ube2D1 and UHRF1 mutants. See Supplemental Figure 3D for the Q70E experiments. Error bars reflect standard deviations from biological duplicates. ** denotes a P-value lower than.01 and *** denotes a P-value below.001.
(D) Rosetta-generated model of the UHRF1-UBL/E2 complex determined using restraints derived from chemical shift perturbations, paramagnetic-spin label distances, and mutations.
(E) Interactions between the residues on the UHRF1-UBL and E2 mutated in panel C. R6 can simultaneously form hydrogen bonds with the D16 sidechain and P17 backbone, residues in the a1-b1 loop (bottom) and Q70 can form hydrogen bonds with the S22 sidechain and the Q20 backbone (top).
To determine the orientation of the UBL domain in the UBL/E2 complex, we introduced a TEMPO paramagnetic spin-label at residues positioned on opposite sides of the UBL β-sheet (UBL residue 9 or 62). NMR-active nuclei in proximity to a paramagnetic species experience line broadening and loss of peak intensity (“PRE,” paramagnetic relaxation enhancement) that is recovered upon reduction of the paramagnetic label. Residues near the N-terminal helix (K8,E9, S11, G24, D28, F31, and H32) of the E2 were most affected when E62C-TEMPO UHRF1-UBL was added to 15N-Ube2D3 (Figure 3B, Supplemental Figure 3B), while D9C-TEMPO UHRF1-UBL affected a distinct set of residues including A19, S22, N41, D42, G47, G48, and S108 (Figure 3B, Supplemental Figure 3C). These data establish a general orientation of the UBL bound to Ube2D that is highly similar to Ub bound to the E2 (see below) (Bosanac et al., 2010, 2011; Brzovic et al., 2006; Buetow et al., 2015).
We used a two-stage approach to generate experimentally-based models for the UHRF1-UBL/Ube2D interaction. First, we used the NMR data described above (chemical shift perturbations and PREs) to perform constrained rigid-body docking with the Rosetta molecular modeling program (see Methods). The resulting models made two specific structural predictions that we tested by mutagenesis. A hydrogen bond network encompassing Ube2D residue S22 and UHRF1-UBL residue Q70 was tested with a Q70E mutation in UHRF1, and a predicted salt bridge between UHRF1-UBL residue R6 and Ube2D residue D16 was tested with charge-swap mutations of R6D in UHRF1 and D16K in Ube2D1. The relevant combinations of E2 and UHRF1 mutants were assayed in H3-peptide ubiquitylation assays (Figure 3C). The activities observed for the mutant proteins support predictions made from the initial models. First, S22R-Ube2D could not function with any of the UHRF1 variants, indicating that backside binding is essential for the activity in the context of the other mutations. Q70E-UHRF1 displayed no activity with any of the three E2 variants (WT, S22R, D16K), consistent with the predicted interaction of the UBL glutamine sidechain with S22 on the E2 (Supplemental Figure 3D). While R6D-UHRF1 displayed no activity with WT-Ube2D, the compensatory D16K-Ube2D mutation rescued H3 ubiquitylation to levels comparable to the WT/D16K-Ube2D pair (Figure 3C). This rescue is particularly striking when considering the R6D-UHRF1 variant has ~6% of the rate of H3ub as the WT. A second stage of modeling was carried out using both the NMR and mutational data, yielding an atomic-level model of the E2/UBL interface in which the UBL position is very similar to that of ubiquitin bound to Ube2D in previously determined experimental structures (Figure 3D, Supplemental Figure 3E, and supplemental file 1). A network of hydrogen bonds at the UHRF1-UBL/E2 interface is revealed: the sidechain of UHRF1-UBL residue R6 interacts with the Ube2D sidechain of D16 and the backbone carbonyl group of P17 (Figure 3E, bottom); the sidechain of UBL residue Q70 interacts with Ube2D residue S22 and the backbone carbonyl on Q20 (Figure 3E, top).
The UHRF1 RING domain (UHRF1-RING) has a lower affinity for E2 than the UBL domain
For most E3 ligases, the RING domain is thought to be the domain responsible for recruiting E2~Ub to form an E2/E3 complex poised for Ub transfer. We were therefore surprised to observe such a profound loss of function by abrogation of the UHRF1-UBL/E2 interaction. Accordingly, we compared the affinities of Ube2D3 for the individual UHRF1-UBL and UHRF1-RING domains. As for the UHRF1-UBL/Ube2D3 complex (see Figure 2), a series of 1H15N-HSQC NMR spectra of either 15N-Ube2D3 or 15N-UHRF1-RING (residues 675–793) were collected with increasing concentrations of unlabeled UHRF1-RING or a stable, isopeptide-linked Ube2D3-Ub conjugate, respectively (Plechanovová et al., 2012). Similar to other RINGs (Metzger et al., 2014), UHRF1-RING recognizes residues in Ube2D3 Helix 1, Loop 4, and Loop 7, a surface distinct from the UBL-binding site defined above (Figure 4A; Supplemental Figure 4A). Analysis of CSPs as a function of the stable Ube2D3-Ub conjugate concentration yielded an apparent Kd of 90 ± 5 μM for binding to 15N-UHRF1-RING. A similar value was obtained for binding to non-conjugated Ube2D3, indicating only a weak preference for E2~Ub conjugate (Supplemental Figure 4B). The analysis shows that the affinity of the UHRF1-RING/Ube2D3-Ub complex is ~6-fold weaker than that of the UHRF1-UBL/E2 interaction (~15 μM). We previously reported the affinity between full-length UHRF1 and E2~Ub to be 3.2 μM (Harrison et al., 2016a) —tighter than either of the individual pairwise interactions.
Figure 4 |. The UHRF1-UBL does not enhance E2~Ub binding to the RING.
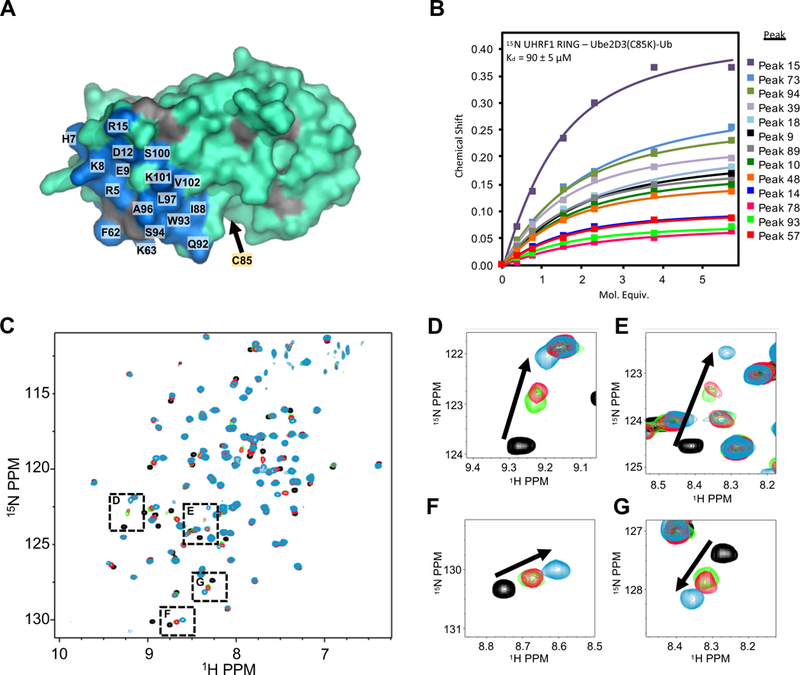
(A) UHRF1-RING binding site (blue on cyan) on the E2 Ube2D3 as determined by 1H15N-HSQC experiments containing 200 μM 15N-Ube2D3(C85S/S22R) and 50 μM UHRF1-RING domain (see methods and Supplemental Figure 4A). Gray; prolines.
(B) Binding curves generated from NMR peak chemical shifts of 82 μM 15N-UHRF1-RING as a function of increasing Ube2D3(C85K/S22R)-Ub. The UHRF1-RING spectrum is not assigned.
(C) 1H15N-HSQC of 100 μM 15N UHRF1-RING in the absence (black) or presence (green) of 100 μM Ube2D3(C85K)-Ub (E2-N-Ub; isopeptide linked E2-Ub) or (red) in the presence of 100 μM E2-N-Ub and 175 μM UHRF1-UBL. The blue spectrum is of 100 μM 15N-UHRF1-RING in the presence of 700 μM S22R-E2-N-Ub (near saturation).
(D-G) Close-up views of peaks indicated in panel C.
The UBL domain does not stimulate UHRF1-RING E3 activity
It has been reported that non-covalent binding of Ub to the backside surface of Ube2D~Ub promotes a closed conjugate conformation, increases the affinity of some RING/E2 interactions, and enhances discharge of Ub (Supplemental Figure 1A) (Buetow et al., 2015). As Ub and UHRF1-UBL bind the same Ube2D surface, we assessed whether UBL binding could promote Ub discharge from Ube2D3~Ub to enhance UHRF1-RING activity. First, we examined whether UHRF1-UBL increases the affinity of Ube2D~Ub for UHRF1-RING. Addition of unlabeled Ube2D3(C85K)-Ub to 15N-UHRF1-RING results in CSPs that exhibit fast exchange behavior, in which peak positions are determined by the relative populations of the free (black peaks in Figures 4C-G) and E2-bound state (blue peaks in Figures 4C-G) of UHRF1-RING. Ube2D(C85K)-Ub was added to 15N-UHRF1-RING at a concentration near the midpoint of the titration trajectory (green spectrum in Figures 4C-G). Following addition of excess UHRF1-UBL, the NMR spectrum was recollected (red spectrum in Figures 4C-G). Only minor perturbation of peaks in the UHRF1-RING spectrum was observed, indicating a minimal change in the fractions of free UHRF1-RING and E2-bound RING. Thus, binding of UHRF1-UBL to the backside of Ube2D only very weakly enhances the interaction between Ube2D3~Ub and the UHRF1-RING in the context of these isolated domains. Consistent with this finding, the presence of UBL in large excess had no detectable effect on conjugate reactivity in aminolysis reactions (Supplemental Figure 4C and D). Hence, UHRF1-UBL binding to the E2 neither enhances the affinity of UHRF1-RING for E2 nor the ubiquitin ligase activity of the isolated UHRF1-RING domain.
Mutations to residues outside the UHRF1-UBL/E2-binding surface disrupt ubiquitylation
Ruling out a Ub-like role of allosterically enhancing E2~Ub discharge, we performed mutational analysis on the UHRF1-UBL to further define its role in UHRF1-mediated ubiquitylation. Three classes of mutations were tested: 1) mutation of residues at or near the UBL-Ube2D interface (R6D, M8R, F46V, R48E, K50R, R64E, Q70E, and L72A), 2) mutation of residues distant from the E2-binding site (W2V, L65A, F59V, H56A, and E62K), and 3) mutations found in COSMIC (Catalogue of Somatic Mutations in Cancer). Consistent with the UBL’s role in E2 recruitment, mutations made directly in the E2-binding interface (R6D, M8R, F46V, R48E, Q70E, and R64E; highlighted in red in Figure 5, Supplemental Figures 5 and 6) substantially reduce the rates of UHRF1-mediated histone H3 and auto-ubiquitylation (Figure 5B and Supplemental Figure 5A) and yield no detectable Ube2D binding by isothermal titration calorimetry (ITC) (Figure 5C and Supplemental Figure 5B). Three of the mutants that lie outside the E2/UBL interface (L72A,H56A, and E62K; colored gray in Figure 5A and B) have little or no effect on ubiquitylation by UHRF1, but unexpectedly, four of the mutants (W2V, L65A, K50R, and F59V) showed defects in H3 ubiquitylation, but not in UHRF1 auto-ubiquitylation (Figure 5A and B and Supplemental Figure 5A) (these residues are shown in green in Figure 5 and Supplemental Figures 5 and 6). These mutants also retain their ability to bind the E2 with similar affinities as WT UHRF1-UBL (Figure 5C and Supplemental Figure 5B). Circular dichroism on a subset of these mutant UBLs confirmed that the proteins are folded, indicating their defects are not a result of misfolding (Supplemental Figure 5C). Defective H3 ubiquitylation with retention of E2 binding and auto-ubiquitylation activity imply a role in substrate specificity for the UBL. The identified residues describe a surface on the UBL potentially involved in intramolecular interactions (Figure 5A, green). Importantly, the loss of activity seen in mutants that disrupt the E2/UBL interaction and in those that have a defect in H3-peptide ubiquitylation (Figure 5B) are also defective in modifying mononucleasomes (Figure 5D). As showed previously, addition of he5mc in trans stimulates the ubiquitylation activity of WT UHRF1 towards nucleosomal H3 in (Figure 5D) (Harrison et al., 2016a). None of the mutants tested (M8R, F46V, W2V, and F59V) ubiquitylate mononucleosomes, indicating that the H3 ubiquitylation defects observed are not an artifact of the minimal H3-peptide substrate (Figure 5D).
Figure 5 |. A surface of the UHRF1-UBL not involved in E2 binding is essential for H3 ubiquitylation.
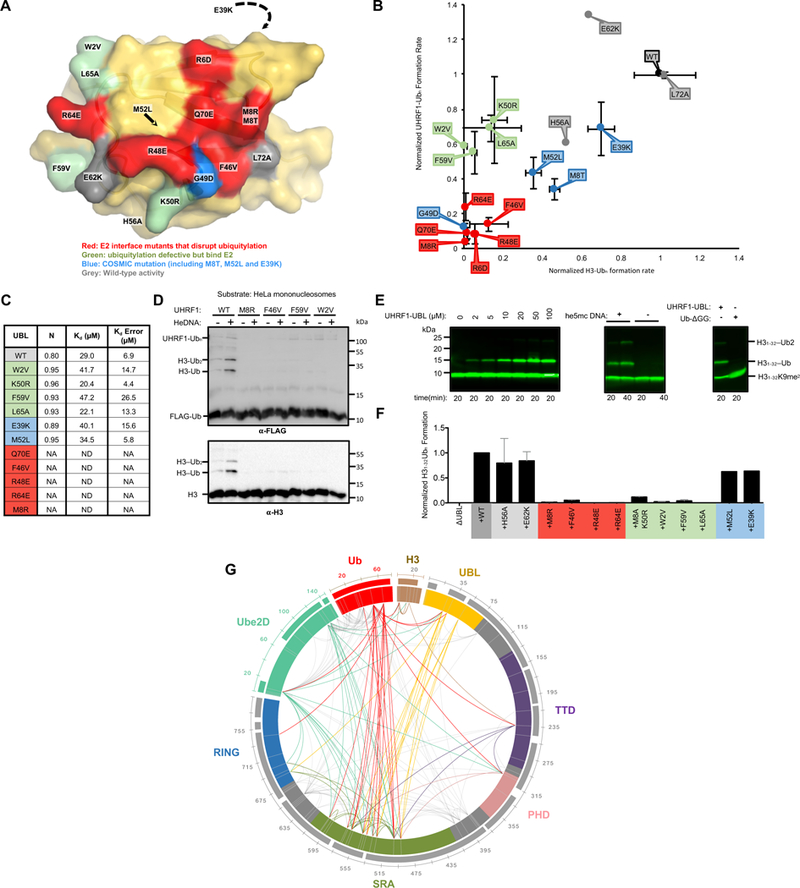
(A) Surface representation of the UBL domain with the mutation sites colored as indicated.
(B) Rates of ubiquitylation (see Methods) for assays containing the indicated mutants of UHRF1. Ubiquitylated substrate (H31–32K9me2; H31–32) and auto-ubiquitylated E3 products are both visualized in this dot plot. Errors reflect the standard deviation from biological duplicates; representative gels from which these rates were determined are in Supplemental Figure 5A.
(C) Kd values for indicated UBL variants binding to Ube2D1 as determined by ITC. Isotherms shown in Supplemental Figure 5B.
(D) HeLa mononucleosome ubiquitylation assay using WT or UHRF1 mutants (see Methods).
(E) H3 peptide (H31–32K9me2; H31–32) ubiquitylation assays in the presence of he5mc-DNA with ΔUBL and increasing UHRF1-UBL added in trans (left). The rescue of ΔUBL in the absence or presence of he5mc-DNA with 100 μM UHRF1-UBL (middle). (Right) ΔUBL rescue in the presence of 400 μM UHRF1-UBL or 400 μM ubiquitin (residues 1–74, Ub-ΔGG). Western blot is for biotin on the H3 peptide.
(F) Densitometry quantification of H3 substrate (H31–32K9me2-biotin) ubiquitylation ΔUBL rescue experiments with the indicated UHRF1-UBL mutants (see Supplemental Figure 6D).
(G) Chemical cross-linking/mass spectrometry analysis of the complex formed between UHRF1, he5mc-DNA, H3-peptide, and isopeptide-linked Ube2D-Ub conjugate. Crosslinks with q-values ≥ 0.01 are shown. Links that involve flexible linker regions of UHRF1 are in gray; those that involve structured domains are color-coded by domain (priority is given to the N-terminal-most domain). Sequence coverage of the MS data is shown as bars outside the circle plot.
Given the apparent importance of the UHRF1-UBL in targeting histone H3, we also tested mutations present in the COSMIC database. Seven of the 99 missense mutations in COSMIC (supplemental file 2) are in the UBL and several of these mutations lie at the E2 interface (Supplemental Figure 5D). We tested four COSMIC UBL mutants (M8T, E39K, G49D, and M52L) for their ability to support histone H3 ubiquitylation and UHRF1 auto-ubiquitylation. All four tested COSMIC mutations show defects in ubiquitylation levels, with the most severe defects observed for the G49D variant located at the E2-interface; M8T (interface), M52L (core) and even E39K result in minor but significant H3 and auto-ubiquitylation defects (Figure 5A and B). Collectively the analysis demonstrates an essential role for the UBL not only in E2 recruitment but also in histone targeting.
The UHRF1-UBL domain can rescue the H3 ubiquitylation defect of the ΔUBL truncation
We speculated that UHRF1-UBL exerts its substrate targeting function via intramolecular interactions within UHRF1. In ubiquitylation assays with the ΔUBL-UHRF1 truncation, both H3 ubiquitylation and auto-ubiquitylation activity were partly restored in a dose-dependent manner when the UBL domain was added in trans (Figure 5E, left and Supplemental Figure 6A). Maximum recovery of H3 ubiquitylation was achieved at 50 μM UBL. Importantly, the UHRF1-UBL rescue of ΔUBL is still dependent on he5mc, underscoring the role of he5mc binding by the SRA domain for H3 ubiquitylation (Figure 5E, center). The recovery is specific to UHRF1-UBL, as neither Ub nor the homologous UBL domains from UHRF2 or Parkin rescue ΔUBL activity, even at high concentrations (400 μM; Figure 5E, right and Supplemental Figure 6B and C). Therefore, the ability of UHRF1-UBL to rescue the ΔUBL-UHRF1 loss of function extends beyond its ability to bind and recruit the E2.
We also tested the ability of mutant UBLs to complement ΔUBL activity. Neither mutations at the E2-binding interface nor near the W2 patch rescued substrate (H31–32K9me2 peptide) ubiquitylation activity when added in trans to ΔUBL (Figure 5F and Supplemental Figure 6D). However, UBL variants capable of interacting with the E2 (W2V, F59V, K50R, and L65A) rescued ΔUBL auto-ubiquitylation activity, while UBL variants unable to bind to the E2 did not (Supplemental Figure 6E).
The UHRF1-UBL facilitates multiple intra- and intermolecular interactions.
Previous studies support a model in which UHRF1 activity is regulated by intramolecular interactions among its domains (Gao et al., 2018; Gelato et al., 2014; Harrison et al., 2016a; Houliston et al., 2017). Although three-dimensional structures have been determined for each domain within UHRF1, there is no experimentally-determined structure of full-length UHRF1. The domains of UHRF1 are connected by long flexible linkers, which may be why full-length UHRF1 has evaded structural determination.
To determine interdomain contacts in UHRF1 that is poised to transfer Ub to its substrate, we performed cross-linking followed by mass spectrometry of the UHRF1/he5mc/H3-peptide complex in the presence of a stable isopeptide-linked E2-Ub conjugate. Upon incubation with bis(sulfosuccinimidyl)suberate (BS3; a lysine-to-lysine cross-linker), extensive inter- and intramolecular crosslinks were observed (Figure 5G and Supplemental Figure 6F). Two incubation time points (10’ and 45’) were analyzed and high confidence filters of q-values ≥ 0.01 were applied to minimize false positives. The data are displayed as a circle plot (Figure 5G). Importantly, numerous crosslinks are observed for previously defined interactions and for interactions we expect to see based on binding experiments in this study. For example, the H3-peptide crosslinks to its binding domain (the TTD), and both the RING and UBL domains of UHRF1 crosslink to both subunits of the Ube2D-Ub conjugate. In addition, many intra- and intermolecular crosslinks are observed for the UHRF1/he5mc/H3-peptide complex. Strikingly, a majority of links emanate from the SRA domain, identifying it as the central hub of the complex: there are links from the SRA to every other structured domain in UHRF1, the E2, Ub, and the H3-peptide. Though much smaller and containing only three lysines, the UBL has the next highest number of inter-domain links, primarily to the SRA and RING domains. Despite good sequence coverage and five lysines, only a few high confidence links were detected from the substrate-binding TTD, which had intramolecular links to the SRA and intermolecular links to the H3 peptide, and Ub. Altogether, the data imply that the UBL and SRA domains are located proximally in the active E3/E2~Ub complex, providing a rationale for the coupling of he5mc binding (to the SRA) and UBL function. Incorporating observations presented above that the UBL recruits the E2, which in turn binds to the RING, we propose that the UBL domain orchestrates hemi-methylated-DNA-dependent targeting of histone H3 through interactions with both the E2 and the SRA.
Other UBLs found in E3 ligases bind to the backside of Ube2D.
Our results show unambiguously that a key role of the UBL domain of UHRF1 is E2 recruitment. This unexpected finding raises the possibility that UBLs in other E3 ligases have similar functions. Several E3s contain N-terminal UBL domains with high sequence homology to UHRF1-UBL, including UHRF2, a homologue of UHRF1 that does not rescue DNA methylation defects in UHRF1 knock-down in cells (Zhang et al., 2011), Parkin, an E3 ligase involved in mitophagy (Harper et al., 2018) and known for mutations in familial cases of Parkinson’s disease (Cruts et al., 2012), and HOIL1, a subunit of the multi-subunit LUBAC E3 complex that attaches linear Ub chains to NFκB and is involved in the innate immune response (Liu et al., 2017) (Figure 6A). NMR binding studies with 15N-Ube2D3 and UBL domains of UHRF2, Parkin, and HOIL1 confirmed that UHRF2 and Parkin UBLs bind to Ube2D as indicated by CSPs in the Ube2D spectrum (Figure 6B and C, left panels)1. The UBL binding surfaces defined by the CSPs map to the backside of Ube2D3 (Figure 6D and E; Supplemental Figure 7A and B). Consistent with this result, no binding was detected by NMR for Ube2D harboring the S22R mutation (Figure 6B and C, right panels). Reciprocal CSP mapping of Ube2D binding to 15N-UHRF2-UBL revealed that UHRF2 uses the same surface of its UBL as UHRF1 uses to interact with Ube2D (Figure 6F; Supplemental Figure 7C). Conservation within the hydrophobic surface of Ub and UHRF1/2 UBLs responsible for E2 binding suggest that the analogous surface of Parkin is also responsible for the observed binding (Figure 6A). Despite this conservation, the UHRF2 and PARKIN UBLs bind to Ube2D3 with considerably lower affinity than the UHRF1-UBL/E2 association, with apparent Kd values of 215 ± 11 μM and 250 ± 11 μM, respectively (Figure 6G and H). These values are much closer to values measured for Ub binding to Ube2D (200–300 μM) (Brzovic et al., 2006; Buetow et al., 2015) than to the UHRF1-UBL binding affinity of ~15 μM measured by NMR in this study. Thus, while UHRF1-UBL stands out for its high-affinity binding to Ube2D, the results reveal a conserved ability of these UBL domains to bind to Ube2D in a manner similar to Ub itself.
Figure 6 |. UBL domains from other E3 ligases interact with Ube2D.
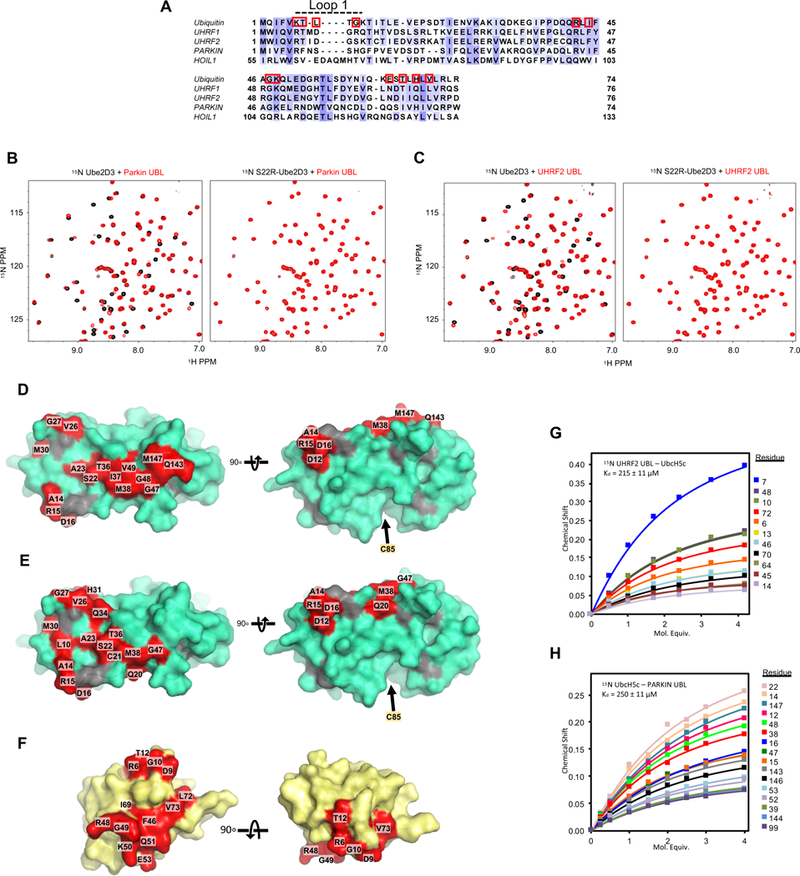
(A) Structure-based multiple sequence alignment of Ub and the UBL domains of UHRF1, UHRF2, Parkin, and HOIL1 are colored by conservation. Red outlines indicate residue side-chains in Ub at the E2/Ub non-covalent interface.
(B and C) 1H15N-HSQC spectrum of 150 μM 15N-Ube2D3(C85S) or 150 μM 15N-Ube2D3(C85S/S22R) in the absence (black) or presence (red) of the indicated UBL: (B) Parkin UBL (C) UHRF2 UBL.
(D and E) Binding sites of Parkin UBL (D) or UHRF2 UBL (E) on the surface of Ube2D3.
(F) Binding site of Ube2D3(C85S) on UHRF2-UBL (PDB 1WY8; BMRB entry 11266) colored red (see methods and Supplemental Figure 7A-C).
(G and H) Binding curves generated from the 1H15N-HSQC peak chemical shifts of 140 μM 15N UHRF2-UBL as a function of Ube2D3(C85S) concentration (G) or 150 μM 15N-Ube2D3 with increasing concentrations of Parkin UBL (H).
Given their conserved E2-binding abilities, we considered the possibility that UHRF1, UHRF2, and Parkin UBLs may enhance the reactivity of the E2~Ub conjugate, similar to reported effects of Ub binding. We recapitulated the observations that non-covalent binding of Ub to Ube2D-O-Ub (C85S oxyester-linked conjugate) enhances E2 discharge mediated by the Ubox domain (functionally equivalent to a RING domain) of the E3 ligase E4B (Supplemental Figure 7D) (Buetow et al., 2015). None of the UBL domains tested (UHRF1-UBL, UHRF2-UBL, or PARKIN-UBL) stimulated this activity (Supplemental Figure 7D). Failure of the E3 UBLs to enhance E2 discharge cannot be due to stronger versus weaker binding, as UHRF1-UBL binds tighter than Ub while the other UBLs bind with similar affinities to Ub. Hence, while UBL binding to Ube2D is a property shared among Ub and the UBLs tested herein, the ability to allosterically enhance E2~Ub reactivity is not. A notable difference in the UBL sequences compared to Ub is in loop 1 (Figure 6A), where each UBL has an insertion of at least one amino acid. This loop of Ub interacts with the α1-β1 loop of Ube2D and is putatively involved in E2~Ub allostery (Buetow et al., 2015), so we speculate that the inability of the UBLs to enhance E2~Ub reactivity arises from divergence in this region. These results provide an impetus for future studies to learn whether E3s that contain UBLs use them as E2-binding modules and what role such binding plays in their ligase function.
DISCUSSION:
Despite the lack of a structure for the full-length enzyme, emerging models based on results from numerous studies define remarkable coordination among the domains that gives rise to UHRF1 function. In the absence of chromatin or other binding partners, UHRF1 exists in a state in which the chromatin-binding domains, TTD/PHD and SRA, interact with each other (Fang et al., 2016; Harrison et al., 2016a) and the polybasic linker that connects the SRA and RING domains binds to the histone-binding site (Gao et al., 2018; Gelato et al., 2014) (Figure 7, left). This conformation provides a physical coupling of the domains responsible for histone and DNA binding and provides a rationale for the positive allostery observed for histone and DNA-binding (Harrison et al., 2016a). The TTD and PHD act together to bind to histone H3 that lacks methylation at Arg2 (Rajakumara et al., 2011) and carries a methyl mark at Lys9 (Rothbart et al., 2012a). Recognition of he5mc-containing DNA by the SRA domain allosterically activates mono-ubiquitylation of histone H3 catalyzed by the RING domain (Harrison et al., 2016a). Our studies define an integral role for the UBL domain in this process. Here we discover that the previously uncharacterized UBL performs two critical functions that ensure histone H3 ubiquitylation by UHRF1. First, despite harboring a RING domain at its C-terminus, the UBL is required to recruit the E2, Ube2D, to UHRF1 (Figure 7). The UBL has a ~3–6-fold higher affinity for E2 than the RING domain and disruption of the UBL/E2 interaction results in a loss of all ubiquitin products formed by UHRF1. Thus, both the UBL and RING domains are responsible for recruiting an E2~Ub to generate an active UHRF1/E2~Ub complex. Second, a separate surface on the UBL is responsible for targeting ubiquitin to histone H3 (Figure 7; right). Disruption of this site specifically diminishes H3 ubiquitylation, leaving auto-ubiquitylation, and perhaps other ubiquitylation activities intact. Our results establish that the UHRF1-UBL acts both to recruit the E2~Ub conjugate and to position this species for direct transfer of ubiquitin to the substrate. Other studies have established the critical role of the UBL in DNA methylation inheritance by demonstrating that a variant lacking the N-terminal UBL domain cannot support DNA methylation in cells (Li et al., 2018; Smets et al., 2017). One study proposed that the UHRF1-UBL can bind and activate DNMT1 (Li et al., 2018) raising the possibility that the UHRF1-UBL not only plays a central role in UHRF1 activity but also that of DNMT1. Alternatively, the in-cell data in that paper could be explained by the H3-ubiquitylation defect we observe for the ΔUBL construct. A related study further implies that the function of the UBL in DNA methylation is through its role in ubiquitylation, showing that UHRF1 with a mutation in UBL residue F46, which we show to be in its E2-binding surface, is not methylation competent (Foster et al. Related Submission). Along with the existing body of literature on UHRF1, the newly defined functional roles for the UBL herein support a model in which all five of the domains in UHRF1, an E2~Ub conjugate, and he5mc-containing DNA coordinate to support H3 ubiquitylation (Figure 7).
Figure 7 |. Model for the bifunctional role of the UBL domain in UHRF1 ubiquitylation of histone H3.
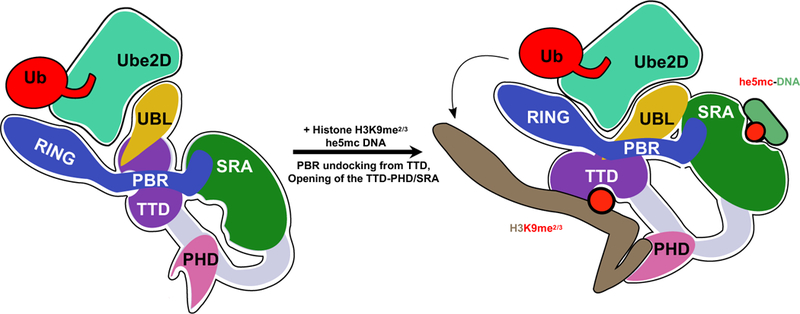
In the absence of chromatin (left), UHRF1 adopts a condensed conformation in which the histone- and DNA-binding modules interact and the linker that connects the SRA and RING (poly-basic region; PBR) occupies the histone-binding site of the TTD. In this state, both the UBL and RING domains can interact with E2~Ub, as judged by ITC and auto-ubiquitylation (Harrison et al., 2016a). Binding to histone or DNA, opens the protein allowing for high-affinity interactions with chromatin. However, productive H3 ubiquitylation is only achieved when he5mc DNA is bound to the SRA (right). The UBL recruits Ube2D, and also positions H3 near the active-site of the E2 through inter-domain interactions with UHRF1. Our cross-linking analysis indicates that the SRA domain is in close proximity to the UBL.
That a small domain with a fold similar to ubiquitin simultaneously plays two distinct functional roles is remarkable. It is not surprising that a domain with high similarity to Ub uses a surface analogous to the well-known “lle-44 hydrophobic patch” of Ub for a protein-protein interaction; this is by far the most highly utilized surface of Ub in its myriad interactions (Harrison et al., 2016b). It is also not surprising that the UBL surface can interact with the “backside” surface of Ube2D family members, as non-covalent binding of Ub to this site was the first of many interactions discovered to involve the E2 backside (Brzovic et al., 2006). Several functional consequences of Ub binding to the backside of an E2 have been reported. The interaction restricts poly-ubiquitin chain formation by Ube2D or by the E2 RAD6 (Brzovic and Klevit, 2006; Hibbert et al., 2011; P. Kumar et al., 2015) and, analogously, SUMO binding to the backside of the SUMO-specific E2, Ubc9, restricts poly-SUMO chain formation (Knipscheer et al., 2007). Ub binding to the Ube2D backside plays an allosteric role in activation of the E2 in concert with some RING-type domains (Buetow et al., 2015). Despite working with the promiscuous chain-building E2, Ube2D, UHRF1 adds mono-ubiquitin to its H3 substrate to recruit DNMT1 (Ishiyama et al., 2017). It is tempting to speculate that binding of UHRF1-UBL to the Ube2D backside may limit poly-ubiquitin chain formation by Ube2D when working with UHRF1.
Our findings add UHRF1 to a growing number of RING E3 ligases that use domains outside the RING domain to interact with the backside surface of their cognate E2s (Brown et al., 2015; Das et al., 2013; Hibbert et al., 2011; Li et al., 2015; Metzger et al., 2013; Turco et al., 2015). In previously reported examples, the accessory E3 domains are α-helical and increase E3 activity by increasing the affinity of the E2/E3 interaction (Das et al., 2013; Hibbert et al., 2011; Li et al., 2015; Metzger et al., 2013). While the UHRF1-UBL domain also increases the ligase’s affinity for its E2, the additional substrate targeting function we have defined for this domain appears to be unique among E3 accessory domains.
Aberrant methylation has long been considered a hallmark of cancer. The global hypomethylation that occurs leads to activation of oncogenes and genome instability. Here we show that UHRF1-UBL mutations observed in cancer genomes disrupt H3 ubiquitylation. Notably, another study found that deletion of the UBL leads to increased cell proliferation (Jenkins et al., 2005), consistent with the biochemical loss of function we report here for DUBL. Collectively the mutations support a key role for the UBL domain in UHRF1 ubiquitylation and imply that disruption of UHRF1 ligase activity may result in disease. Indeed, there are also 11 UHRF1-RING mutations in COSMIC (supplemental file 2), providing further evidence for connections between UHRF1 Ub ligase activity, the dysregulation of methylation patterns, and genome stability. However, while a link between UHRF1 and cancer appears strong, the relevant functions and dysfunctions of UHRF1 associated with disease are likely to be multifaceted.
STAR METHODS:
Protein Expression and Purification
All UHRF1 and UHRF2 truncations and mutations were cloned into a modified version of the pQE vector containing a Tobacco Etch Virus (TEV) protease cleavable His6-MBP tag (Harrison et al., 2016a). Parkin-UBL (residues 1–76) was cloned into a pCool vector (gift of N. Zheng) with an N-terminal Glutathione S-transferase tag (GST tag) and TEV cleavage site using ligation-independent cloning. UHRF1 constructs including full-length (residues 1–793), DUBL (residues 125–793), UHRF1-UBL (residues 1–76), and UHRF1-RING (residues 675–793), UHRF2-UBL (residues 1–81), and Parkin-UBL (residues 1–76) and mutants thereof were generated using a combination of standard PCR based site-directed mutagenesis, restriction cloning, and/or with Gibson Assembly (Synthetic Genomics, Inc.). All proteins were expressed in Escherichia coli (E. coli) grown to O.D. 0.6–0.8 at 600 nm, induced with 0.6 mM IPtG, and grown overnight at 16–18 °C. Cultures were centrifuged at 5000 rpms for 15 min and the cell pellet was resuspended in 25 mM Tris pH 7.6, 200 mM NaCl for cell lysis and subsequent protein purification. Phenylmethylsulfonyl fluoride (PMSF; Sigma, Cat# 329–98-6) was added to a final concentration 1 mM to the resuspended cell pellet immediately prior to cell lyses, which was performed using a short sonication, followed by homogenization or by two passages through a French pressure cell. Insoluble debris was pelleted by centrifugation at 17,500 rpm for 30 min to 1 hour at 4 °C. UHRF1 and UHRF2 variants were passed over Ni-NTA resin (Clonetech, Cat #635660), washed, eluted with imidazole, and cleaved with TEV protease (purified according to previous literature). The cleaved MBP-tag was then removed by anion exchange with a HiTRAP Q-HP column (GE Healthcare, Cat #17115401) using a gradient elution from 50–250 mM NaCl in a Tris pH 8.0 buffer. When removing the MBP tag from the isolated UBL domains and mutants, the UBL domain did not bind to the column at 50 mM NaCl. Alternatively, the His6-MBP tag was removed with a combination of recapturing the tag on Ni-NTA resin after dialysis into 25 mM Tris-HCl pH 7.6, 200 mM NaCl, and 5 mM imidazole, and passage through amylose resin (New England BioLabs, cat# E8021S). Full-length UHRF1 and mutants, DUBL, and all proteins used for NMR experiments were further purified by size exclusion chromatography (SEC) with a Superdex 200 HiLoad 16/600 column (GE Healthcare, Cat #28989335), a Superdex 200 10/300 GL column (GE Healthcare Cat #17517501), or a Superdex 75 10/300 GL column (GE Healthcare, Cat# 17517401). GST-Parkin-UBL was purified using glutathione Sepharose 4B resin (GE Healthcare, Cat# 17075601), followed by cleavage of the GST-tag with TEV protease, dialysis, removal of GST using glutathione Sepharose resin, and SEC using a Superdex 75 10/300 GL column. E4B Ubox was expressed in with an N-terminal His6-SUMO tag followed by a H3C protease cleavage site, and purified as previously described (E4BU, residues 1105–1186) (Nordquist et al., 2010). Ubiquitin-conjugating enzymes were purified with various affinity purification techniques: Ube2D1, Ube2D2, Ube2D3 (and mutants thereof), Ube2E1, Ube2E2, Ube2E3, Ube2L3, Ube2W, Mms2 (Ube2V1), Ube2G2, Ube2I, Ube2S2, Ube2J2 (catalytic domain), Ube2H, Ube2T, Ube2V2, and Hisa-Ube2N were purified as previously described.
H3-peptide ubiquitylation assays
A typical ubiquitylation assay contained 1.5 μM full-length or mutant UHRF1, 0.5–0.05 μM human E1, purified in house or purchased (BostonBiochem, Cat #E-304), 5–0.5 μM of the indicated E2, 6–20μM of either HA-tagged or FLAG-tagged Ub, purified in-house or purchased (Boston Biochem, Cat #U-120), 2.5 mM MgCl2, 5 mM ATP, and unless otherwise indicated, 4 μM hemi-methylated DNA (a 12 base-pair duplex DNA with one strand containing a single 5-methylcytosine (5mC); Sequence: CCATG5mCGCTGAC; IDT DNA). H3(1_32) peptides with a C-terminal biotin tag were used and the peptides were synthesized as previously described (Rothbart et al., 2012b). Methodological differences are the results of performing similar experiments in separate labs and the corroboration of the data even when using different sets of reagents underlie the robustness of the results.
The time-course assays were performed as stated above, except TAMRA-labeled ubiquitin was used, allowing for fluorescent visualization of the SDS-page gels immediately after running. These assays contained mutants or WT UHRF1, 3 μM MBP-tagged E3 was used and the tag was cleaved with TEV immediately prior to the ubiquitylation reaction. We have observed decreased activity of N-terminally tagged UHRF1 (data not shown). Most time course assays were repeated in duplicate (except E62K and H56A), but single time point assays with each mutant was replicated between 3–5 times with consistent results. Apparent Initial rates were determined through densitometry quantification of fluorescent H3ub bands using the GelAnaylzyer software and fitting the initial phase of the data to a linear equation. Hela mononucleosomes were purchased commercial (EpiCypher 16–0009) and 500 nM was added to the reactions with the indicated UHRF1 variants.
For ΔUBL rescue experiments, UHRF1-UBL variants were added in trans at the indicated concentrations, all the mutants were added at 100 μM. Time zero samples were collected before the addition of ATP. Reactions were quenched with SDS-PAGE sample buffer and analyzed by gel electrophoresis followed by immunoblotting blotting for HA (mouse anti-HA monoclonal antibody, Invitrogen, Ref#: 26183; Alexa Fluor® 680 goat anti-mouse IgG,
Molecular Probes, Ref # A21058) or FLAG (BioLegends 637303; Abcam Goat Anti-Rat IgG H&L Alexa Fluor® 647 ab150159), and/or biotin (Alexa Fluor® 790 Streptavidin, Jackson ImmunoResearch, Ref# 016–650-084 or AlexaFluor® 488 Streptavidin Fisher Ref# S32354).
NMR spectroscopy
All 15N-labeled proteins were produced in E. coli using MOPS minimal media supplemented with 15N-ammonium chloride (Cambridge Isotope Laboratories Incorporated, Item# NLM-467). Bacterial cultures were grown to an optical density of 0.6–1.0 at 600 nm at 37 °C, induced with 400 jM IPTG, and grown overnight for about 16 hours at 16 °C. For doubly labeled 15N,13C UHRF1-UBL, cultures were grown and expressed as above in the presence of 2 g/L of 13C-D-glucose (Sigma Aldrich, Product# 389374). After overnight expression cells were harvested and proteins were purified the same way as their unlabeled counterparts.
Unlabeled or 15N-labeled Ube2D3(C85K)-Ub or Ube2D3(S22R/C85K)-Ub (isopeptide linked conjugate) was generated by combining 400 μM Ube2D3(C85K) or Ube2D3(S22R/C85K), 8.4 μM E1, 5 mM ATP, 10 mM MgCl2, and 600 μM Ub in a reaction buffer of 25 mM sodium phosphate pH 7.0 and 150 mM NaCl. Immediately after addition of ATP the sample was diluted to twice the volume using a high pH Tris buffer composed of 50 mM Tris pH 10.0 and 150 mM NaCl. The reaction was incubated for 16 hours overnight at 37 °C and purified by SEC in 25 mM sodium phosphate pH 7.0 and 150 mM NaCl for NMR experiments. Unlabeled or 15N-labeled Ube2D3(C85S) or Ube2D3(S22R/C85S) conjugate was generated as described above.
All NMR data were processed with the NMRpipe package and visualized in NMRviewJ; peak intensities and chemical shift perturbations (CSPs) where measured using NMRviewJ using the formula Δδj = [(15NΔδj/5)2 + (1HΔδj)2]1/2, where 15NΔδj and 1HΔδj represent the difference in chemical shift between the bound and unbound states for 15N and 1H chemical shifts, respectively.
To assign the UHRF1-UBL we used a standard set of multidimensional NMR experiments (HNCA, HNCOCA, HNCACB, CBCACONH) collected on an Avance III 600 MHz spectrometer equipped with a cryoprobe. Triple resonance experiments were performed with 760 μM 15N13C UhRf1-UBL (residues 1–76) using a construct that contained short, non-native N- and C-terminal extensions: GSSRVD and KLN, respectively, at 25 °C buffered at pH 7.0 (in 25 mM sodium phosphate pH 7.0, 150 mM NaCl, 1 mM DTT) in the presence of 10% D2O. Resonance assignments were performed by correlating peaks among the triple resonance experiments in NMRviewJ using known variations in backbone amide proton, nitrogen atoms, and CαCβ chemical shifts and known secondary structural elements within the crystal structure of UHRF1-UBL (PDB code 2FAZ) to guide and verify assignments with the aid of Alex Maltsev’s SBiNLab java script and published calculations (Kjaergaard and Poulsen, 2011; Kjaergaard et al., 2011).
All 1H15N-HSQC and 1H15N-HSQC-TROSY experiments were collected on either an Avance III 500 MHz NMR, Avance III 600 MHz NMR equipped with a cryoprobe, or an Avance 800 MHz NMR spectrometer with a cryoprobe (Bruker) at the indicated concentrations. Binding experiments of 15N-E2s to UBLs were all performed with 150 μM Ube2D3(S22R/C85S) or Ube2D3(C85S) with and without a 1 molar equivalent of purified unlabeled UBL domain unless otherwise indicated. Reciprocal experiments were performed with 15N-UBL domains and the same calculations. For binding interactions in which Kd values where determined, NMR titration curves were generated with NMRviewJ (version 9.2.0-b11) by plotting chemical shifts as a function of unlabeled protein concentration. All shown individual peak shifts where fit to a quadratic base 10 model with 250 simulations using the group fit option for Kd determination in NMRviewJ. The error generated by NMRviewJ reflects the deviation of the experimental data points from the curve fit, not experimental error. All binding is assumed to fit a 1:1 protein:ligand model. The affinity of the Ube2D3 for UHRF1-UBL was determined using 150 μM 15N-Ube2D3(C85S) and increasing unlabeled UHRF1-UBL. The affinity of UHRF1-RING for the Ube2D3~Ub conjugate was determined using 82 μM 15N-UHRF1-RiNG, for which we do not have assignments, and increasing unlabeled Ube2D3(S22R/C85K)-Ub, while the affinity for the free E2 Ube2D3 was measured using 200 μM 15N-Ube2D3(S22R/C85S) and increasing unlabeled UHRF1-RING domain. The Ube2D3 interaction with UHRF2 UbL was measured using 15N-UHRF2 UBL protein and monitoring its peak shifts with the addition of increasing wild type Ube2D3. The Parkin UBL interaction with Ube2D3 was measured using 150 μM 15N-Ube2D3 and increasing concentrations of unlabeled PARKIN UBL domain.
For all binding interactions, the contact surfaces of each species are shown on a previously determine structure of that protein (or closely related protein), as indicated. Unless otherwise indicated, the surface of PDB 4V3L is shown. To determine the surfaces both CSP and peak broadening was used as criteria for individual residues to be highlighted. Peaks with CSPs greater than one standard deviation from the mean peak CSP were considered significantly perturbed. Analogously, peaks that experience intensity loss greater than one standard deviation from the average intensity loss across all peaks are highlighted as significantly perturbed. In some cases, such as the UHRF1-UBL interaction with Ube2D, some peaks experience intermediate exchange kinetics and their trajectory/shifts could not be tracked even if they reappeared at high concentrations of binding partner. These peaks are included in the peaks significantly perturbed because they experience significant intensity loss.
Spin label experiments for UBL-E2 binding were performed with the paramagnetic spin label, TEMPO. The cysteine-reactive free radical 4-(2-Iodoacetamido)-TEMPO (Sigma-Aldrich, Cat# 253367) was conjugated to UHRF1-UBL mutants D9C or E62C. Conjugation was performed by incubating ~80 μM UHRF1-UBL mutants with 800 μM 4-(2-Iodoacetamido)-TEMPO (from a 100 mM stock solution in DMSO) in 25 mM sodium phosphate pH 7.0 and 150 mM NaCl buffer overnight at room temperature in the dark. The next morning samples were buffer exchanged to remove excess unreacted spin label and concentrated for use in NMR experiments. 1H15N-HSQC-TROSY experiments were performed of 150 μM 15N-Ube2D3(C85S) and unlabeled 150 μM UHRF1-UBL-TEMPO conjugates in the absence or presence of 10 mM ascorbate. Intensity loss due to spin-label effects were determined using the ascorbate containing samples as a reference. The top 5 % of affected peaks are plotted on the surface of the E2 structure (E2 in PDB 4V3L).
Yeast 2-hybrid screen
A yeast 2-hybrid (Y2H) E2 screen was performed with a previous set of E2s cloned into pACT2 vectors (Christensen et al., 2007). The UHRF1-UBL (residues 1–76) was cloned into a pGBD120 (a.k.a. pGBD-C1) vector as bait. Individual E2 and UBL vectors were co-transformed into yeast strain PJ69–4A (RGY1109) and positive transformants were grown and selected on synthetic defined (SD) -Leu -Trp plates. Single colonies were selected and resuspended in 100 pL of sterile water, serially diluted (10-fold dilutions in water) and spotted on the surface of selective SD medium (-His -Trp -Leu; to see Y2H interaction) or nonselective growth media (for spotting efficiency). Yeast were grown for 8 days before being photographed. Positive controls for Y2H were previously verified interacting pairs of pGBD120 San1 and a pGAD-C1 San1 substrate.
E2~Ub aminolysis reactions and oxyester conjugate hydrolysis assays
For aminolysis reactions, wild-type Ube2D3~Ub was generated as previously described (DaRosa et al., 2015). In brief, a reaction mixture containing 100 μM Ube2D3, 5 μM human ubiquitin E1, 200 μM Ub, 2.5 mM MgCl2, and 5 mM ATP was incubated at 35 °C for about 30 mins in a reaction buffer (25 mM sodium phosphate pH 7.0 and 150 mM NaCl) to generate thioester conjugate E2~Ub. The E2~Ub species was purified from this reaction mixture by SEC using a Superdex 75 10/300 GL column in reaction buffer. Freshly purified Ube2D3~Ub was concentrated and used directly for aminolysis reactions.
Purified Ube2D3~Ub conjugate, was mixed with UHRF1-RING domain (residues 685–793) for a final concentration of 25 μM E2~Ub, and increasing concentrations of UHRF1-RING domain, and, where indicated, 150 mM UHRF1-UBL (residues 1–76) in reaction buffer. Zerominute time points were taken prior to the addition of the amino acid lysine (in 500 mM stock solution in reaction buffer) to a final concentration of 40 mM lysine. Time points were collected at 5, 10, and 15 minutes after the addition of lysine and quenched in non-reducing SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE and stained with coomassie-blue.
Ube2D3(C85S)-Ub oxyester conjugate hydrolysis reactions were performed similarly to the reactions above, but in the absence of lysine, as previously reported (Buetow et al., 2015). Oxyester-linked E2-Ub was generated as previously described (Pruneda et al., 2011): a mixture containing 400 μM Ube2D3(C85S) was incubated with 650 μM Ub, 6 μM human E1, 5 μM ATP, and 10 mM MgCl2 at 37 °C for 4 hours in reaction buffer followed by purification by SEC using a superdex 75 10/300 GL column in reaction buffer. Hydrolysis experiments were performed using purified oxyester-linked E2-Ub in a reaction mixture containing 15 μM E2-Ub and 4 μM E4B-Ubox (E4BU; residues 1092–1173) and the indicated amount of Ub(1–74) (residues 1–74) or UBL domain. When performing discharge assays with UHRF1-UBL or UHRF2-UBL at concentrations above 200 μM precipitation was observed. The first time point was collected at 0 (before addition of E4BU) or at 2 mins after the addition of E4BU. Each time point sample was collected and quenched in SDS-PAGE sample buffer. Samples were analyzed using SDS-PAGE and stained with coomassie-blue.
Isothermal Calorimetry
Isothermal Calorimetry (ITC) experiments were performed at 25C with a MicroCal PEAQ-ITC automated calorimeter in 25 mM Hepes pH 7.4, 100 mM NaCl. Most experiments were performed with 100 μM of each UBL titrated with 1mM Ube2D1. Although the assay the WT and K50R had 2mM Ube2D1 and the assay with M8R had 50 μM and 500 μM Ube2D1. Data were fit to a single-site binding model using the Malvern software package.
Circular Dichroism
Circular Dichroism (CD) spectra were collected on a JASCO J-815 CD spectropolarimeter. The UBLs were diluted to 20 μM into CD buffer (25 mM HEPES pH 7.5 and NaCl 100 mM). CD spectra were acquired over a wavelength range of 250–200 nm with a data pitch of 0.5 nm and with a scanning speed of 10 nm/min. To overcome the high absorbance of the buffer the bandwidth was set to 15 nm.
Rosetta Modeling of the UBL/E2 complex
Structures of the UHRF1-UBL domain (PDB code: 2FAZ) and Ube2D3 (PDB code: 5EGG were prepared for Rosetta docking (Chaudhury et al., 2011) by relaxing the structures individually and then combining them into a single file with the UBL positioned ~8A off the backside surface of the E2. Docking then commenced by spinning the UBL domain freely and using a rigid body translation to slide the UBL onto the E2. This was followed by energy optimization with Monte Carlo sampling of rigid body orientation and side chain rotamers as specified by the DockingProtocol mover in Rosetta. Hundreds of thousands of independent trajectories were performed to search for low energy models. Initially, constraints based on the NMR data were implemented as AMBIGUOUS AtomPair constraints in Rosetta using the BOUNDED functional form that penalizes models if the specified atom pairs are not found within the indicated distance range (see constraint files). However, this approach did not converge to a narrow set of docked poses, but rather generated a diverse set of models similar in energy. These results led us to perform the spin-label experiments to experimentally determine the orientation of the UBL relative to the E2.
We incorporated the spin-label information to the modeling in two ways: they were added as AtomPair constraints using the BOUNDED function and they were also implemented as filters to remove structures that did not satisfy the distance requirements. The filtering was applied after the first step of docking with a reduced representation of the amino acid side chains (see Rosetta Script). Using this set of conditions, we produced 200,000 models and on the top 10 models performed 2,000 refinement trajectories with the FastRelax application. The relax application allows limited backbone motion, and in these models we observed several extensive hydrogen bond networks between the UBL and the E2, including an interaction between U1-R6 and Ube2D-D16, an interaction that we validated with mutations (Figure 3C).
Based on the mutational information, we added an AMBIGUOUS constraint between all pairs of the sidechain polar atoms between U1-R6 and Ube2D-D16. We also constructed a new starting pose, by first aligning the UBL to the ubiquitin position in the 4v3l Ub/Ube2D co-crystal structure. Next, to accommodate the interaction between U1-R6 and Ube2D-D16, we performed loop modeling of the α1-β1 loop, using the loop model application in full atom mode. We filtered out loop orientations that dramatically altered the position of the loop. The docking protocol was run on the top 5 scoring loops, followed by relax. We filtered out structures where U1-R6 was not contacting Ube2D-D16. We also observed two possible rotamers for Q70, one where the Q70 forms hydrogen bonds with S22 and Q20, or sometimes P18, and another where Q70 did not appear to contact the E2. The hydrogen bond networks shown in our model were observed in the majority of the low scoring structures that passed these criteria. We did not observe a hydrogen bond between G49, the Ub-A46 homolog, and S22, which was observed in the 4v3l structure (Buetow et al., 2015). However, backbone flexibility was not explicitly allowed in this region, therefore a more extensive hydrogen bond network may exist. See Supplemental file 1 for the RosettaScript, flags, and constraints used.
Chemical Cross-linking and Mass Spectrometry Analysis (XL-MS)
Chemical cross-linking and mass spectrometry analysis (XL-MS) was carried out as described by Zelter et al., 2015 (Zelter et al., 2015). Reactions were 105 μL total volume in 25 mM HEPES pH 7.4, 90 mM NaCl, 1 mM DTT containing 2.1 μM UHRF1, 4 μM H3-peptide (H31–32K9me2-biotin) 10 μM HeDNA, 20 μM E2-Ub plus 1.8 mM BS3. Crosslinking was carried out for 10 mins or 45 mins at room temperature before quenching 35 μL aliquots by addition of 5 μL 1 M NH4HCO3 plus 1 μL 0.5 M EDTA. After quenching, reactions were brought up to 0.1% PPS silent surfactant (Expedion Inc. San Diego, CA) and 5 mM TCEP (tris(2-carboxyethyl)phosphine) and reduced for 60 mins at 60°C. Alkylation was performed at room temperature with 6 mM iodoacetamide, followed by trypsin digestion at 37°C for 6 hours in an Eppendorf Thermomixer with shaking (1000 rpm) at a substrate to enzyme ratio of 15:1 prior to acidification with 250 mM HCl (final concentration). Digested samples were stored at −80°C until analysis. Mass spectrometry and data analysis were performed as previously described (Zelter et al., 2015). In brief, 0.45 μg of digested protein was loaded by autosampler onto a 150-μm Kasil fritted trap packed with 2 cm of Reprosil-Pur C18-AQ (3-μm bead diameter, Dr. Maisch) at a flow rate of 2 μl per min. After desalting with 8 μl of 0.1 % formic acid plus 2% acetonitrile, the trap was brought online with a fused-silica capillary tip column (75-μm i.d.) packed with 30 cm of Reprosil-Pur C18-AQ (3-μm bead diameter, Dr. Maisch). Peptides were eluted from the column at 0.25 μL/min using an acetonitrile gradient. A QExactive HF (Thermo Fisher Scientific) was used to perform mass spectrometry in data dependent mode. Acquired spectra were converted into mzML using msconvert ProteoWizard (Chambers et al., 2012). All proteins in the sample were identified using Comet (Eng et al., 2013). Cross-linked peptides were identified within proteins identified by comet, using Kojak version 1.4.3 (Hoopmann et al., 2015) available at (http://www.kojak-ms.org). A statistically meaningful q-value was assigned to each peptide spectrum match (PSM) through analysis of the target and decoy PSM distributions using Percolator version 2.08 (Kail et al., 2007). Target databases consisted of all proteins identified in the sample analyzed. Decoy databases consisted of the corresponding set of reversed protein sequences. Data were filtered to show hits to the target proteins that had a Percolator assigned peptide level q value ≤ 0.01 and were identified by 2 or more PSMs. The complete unfiltered list of all PSMs and their Percolator assigned q values are available on the ProXL web application (Riffle et al., 2016) at: https://proxl.yeastrc.org/proxl/viewProject.do?project_id=57 along with the raw MS spectra and search parameters used.
Supplementary Material
Table of missense mutations in UHRF1 found in COSMIC, Related to Figure 5
Models, constraints, command line options, and Rosetta script, Related to Figure 3
ACKNOWLEDGMENTS:
This work was supported by grants from the National Institutes of Health: NIH R01GM088055 (REK); NIH T32 GM007270 (PAD), NIH P41 GM103533 (AZ and TND), NIH R01 GM098503 (PSB); and NIH R01GM073960 (Bk). We thank Richard Gardner for help and reagents; Wenqing Xu for advice; the Yeast Resource Center at the UW for resources; Brian Strahl and Krzysztof Krajewski for providing H3 peptides; Nick Brown for providing TAMRA-labeled ubiquitin; Lisa Tuttle for help on NMR assignments.
Footnotes
DECLARATIONS OF INTERESTS:
The authors declare no competing interests.
Indications of E2 binding were observed for HOIL UBL, but the protein was prone to selfassociationunder NMR conditions, limiting a full binding analysis (data not shown).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, and Shirakawa M (2008). Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, and Dhe-Paganon S (2008). Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, et al. (2010). Ubiquitin Binding to A20 ZnF4 Is Required for Modulation of NF-kB Signaling. Mol. Cell 40, 548–557. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Phu L, Pan B, Zilberleyb I, Maurer B, Dixit VM, Hymowitz SG, and Kirkpatrick DS (2011). Modulation of K11-linkage formation by variable loop residues within UbcH5A. J. Mol. Biol. 408, 420–431. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve P-O, Clark A, Pradhan S, and Jacobsen SE (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. [DOI] [PubMed] [Google Scholar]
- Brown NG, VanderLinden R, Watson ER, Qiao R, Grace CRR, Yamaguchi M, Weissmann F, Frye JJ, Dube P, Ei Cho S, et al. (2015). RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc.Natl. Acad. Sci. 112, 5272–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, and Klevit RE (2006). Ubiquitin Transfer from the E2 Perspective: Why is UbcH5 So Promiscuous? Cell Cycle 5, 2867–2873. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, and Klevit RE (2006). A UbcH5/Ubiquitin Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination. Mol. Cell 21, 873–880. [DOI] [PubMed] [Google Scholar]
- Buetow L, Gabrielsen M, Anthony NG, Dou H, Patel A, Aitkenhead H, Sibbet GJ, Smith BO, and Huang DT (2015). Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol. Cell 58, 297–310. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, and Gray JJ (2011). Benchmarking and Analysis of Protein Docking Performance in Rosetta v3.2. PLoS One 6, e22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, and Klevit RE (2007). E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948. [DOI] [PubMed] [Google Scholar]
- Cruts M, Theuns J, and Van Broeckhoven C (2012). Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 33, 1340–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, and Xu W (2015). Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Liang Y-H, Mariano J, Li J, Huang T, King A, Tarasov SG, Weissman AM, Ji X, and Byrd RA (2013). Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J. 32, 2504–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, and Huang DT (2012). BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, and Gehrke C (1982). Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 10, 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, and Hoopmann MR (2013). Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24. [DOI] [PubMed] [Google Scholar]
- Fang J, Cheng J, Wang J, Zhang Q, Liu M, Gong R, Wang P, Zhang X, Feng Y, Lan W, et al. (2016). Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 7, 11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BM, Stolz P, Mulholland CB, Montoya A, Kramer H, Bultmann S, and Bartke T Critical role of the UBL-domain in stimulating the E3 ubiquitin ligase activity of UHRF1 towards chromatin. Mol. Cell. Related Submission [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Tan X-F, Zhang S, Wu T, Zhang Z-M, Ai H, and Song J (2018). An Intramolecular Interaction of UHRF1 Reveals Dual Control for Its Histone Association. Structure 26, 304–311.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelato KA, Tauber M, Ong MS, Winter S, Hiragami-Hamada K, Sindlinger J, Lemak A, Bultsma Y, Houliston S, Schwarzer D, et al. (2014). Accessibility of Different Histone H3-Binding Domains of UHRF1 Is Allosterically Regulated by Phosphatidylinositol 5-Phosphate.Mol. Cell 54, 905–919. [DOI] [PubMed] [Google Scholar]
- Goyal R, Reinhardt R, and Jeltsch A (2006). Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic Acids Res. 34, 1182–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Ordureau A, and Heo J-M (2018). Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93–108. [DOI] [PubMed] [Google Scholar]
- Harrison JS, Jacobs TM, Houlihan K, Van Doorslaer K, and Kuhlman B (2015). Data in support of UbSRD: The Ubiquitin Structural Relational Database. Data Br. 5, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JS, Cornett EM, Goldfarb D, Darosa PA, Li ZM, Yan F, Dickson BM, Guo H, Cantu DV, Kaustov L, et al. (2016a). Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JS, Jacobs TM, Houlihan K, Van Doorslaer K, and Kuhlman B (2016b).UbSRD: The Ubiquitin Structural Relational Database. J. Mol. Biol. 428, 679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, and Cheng X (2008). The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, and Cartron P-F (2018). Specific or not specific recruitment of DNMTs for dNa methylation, an epigenetic dilemma. Clin. Epigenetics 10, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert RG, Huang A, Boelens R, and Sixma TK (2011). E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. U. S. A. 108, 5590–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopmann MR, Zelter A, Johnson RS, Riffle M, MacCoss MJ, Davis TN, and Moritz RL (2015). Kojak: efficient analysis of chemically cross-linked protein complexes. J. Proteome Res. 14, 2190–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houliston RS, Lemak A, Iqbal A, Ivanochko D, Duan S, Kaustov L, Ong MS, Fan L, Senisterra G, Brown PJ, et al. (2017). Conformational dynamics of the TTD-PHD histone reader module of the UHRF1 epigenetic regulator reveals multiple histone-binding states, allosteric regulation, and druggability. J. Biol. Chem. 292, 20947–20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, et al. (2017). Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance. Mol. Cell 68, 350–360.e7. [DOI] [PubMed] [Google Scholar]
- Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang, Huang J., Yam GC, et al. (2005). Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol. Biol. Cell 16, 5621–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kail L, Canterbury JD, Weston J, Noble WS, and MacCoss MJ (2007). Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925. [DOI] [PubMed] [Google Scholar]
- Kjaergaard M, and Poulsen FM (2011). Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 50, 157–165. [DOI] [PubMed] [Google Scholar]
- Kjaergaard M, Brander S, and Poulsen FM (2011). Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR 49, 139–149. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, and Sixma TK (2007). Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 26, 2797–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Magala P, Geiger-Schuller KR, Majumdar A, Tolman JR, and Wolberger C (2015). Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity. Nucleic Acids Res. 43, 9039–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liang Y-H, Mariano J, Metzger MB, Stringer DK, Hristova VA, Li J, Randazzo PA, Tsai YC, Ji X, et al. (2015). Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B. J. Biol. Chem. 290, 30225–30239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wang L, Du Y, Xie S, Yang X, Lian F, Zhou Z, and Qian C (2018). Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Y, Gong Y, Fu T, Hu S, Zhou Z, and Pan L (2017). Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly. Cell Rep. 21, 27–36. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Liang Y-H, Das R, Mariano J, Li S, Li J, Kostova Z, Byrd RA, Ji X, and Weissman AM (2013). A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms. Mol. Cell 50, 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, and Weissman AM (2014). RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K,Shimamura S, Arita K, Kodama T, Ishikawa F, et al. (2013). Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 502, 249–253. [DOI] [PubMed] [Google Scholar]
- Nordquist KA, Dimitrova YN, Brzovic PS, Ridenour WB, Munro KA, Soss SE,Caprioli RM, Klevit RE, and Chazin WJ (2010). Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry 49, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, and Hay RT (2012). Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, and Klevit RE (2011). Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme~ubiquitin conjugate. Biochemistry 50, 1624–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, and Klevit RE (2012). Structure of an E3:E2~Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box Ligases. Mol. Cell 47, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Wolf P, Liu N, Link S, Smets M, Mastra F La, Forne I., Pichler G, Horl D, Fellinger K, et al. (2015). DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 25, 911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. (2011). PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol. Cell 43, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffle M, Jaschob D, Zelter A, and Davis TN (2016). ProXL (Protein Cross-Linking Database): A Platform for Analysis, Visualization, and Sharing of Protein Cross-Linking Mass Spectrometry Data. J. Proteome Res. 15, 2863–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al. (2011). Disorder Targets Misorder in Nuclear Quality Control Degradation: A Disordered Ubiquitin Ligase Directly Recognizes Its Misfolded Substrates. Mol. Cell 41, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. (2012a). Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Strahl BD, and Fuchs SM (2012b). Peptide microarrays to interrogate the “histone code”. Methods Enzymol. 512, 107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith H, and Strahl BD (2013). Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 27, 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. [DOI] [PubMed] [Google Scholar]
- Smets M, Link S, Wolf P, Schneider K, Solis V, Ryan J, Meilinger D, Qin W, and Leonhardt H (2017). DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation. Hum. Mol. Genet. 26, 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, and Meissner A (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220. [DOI] [PubMed] [Google Scholar]
- Turco E, Gallego LD, Schneider M, and Köhler A (2015). Monoubiquitination of histone H2B is intrinsic to the Bre1 RING domain-Rad6 interaction and augmented by a second Rad6-binding site on Bre1. J. Biol. Chem. 290, 5298–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget J, and Mayor T (2010). The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell. [DOI] [PubMed] [Google Scholar]
- Zelter A, Bonomi M, Kim J. ook, Umbreit NT, Hoopmann MR, Johnson R, Riffle M, Jaschob D, MacCoss MJ, Moritz RL, et al. (2015). The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nat. Commun. 6, 8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W, Li J, Dong S, Koseki H, and Wong J (2011). S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 21, 1723–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of missense mutations in UHRF1 found in COSMIC, Related to Figure 5
Models, constraints, command line options, and Rosetta script, Related to Figure 3


