Figure 5 |. A surface of the UHRF1-UBL not involved in E2 binding is essential for H3 ubiquitylation.
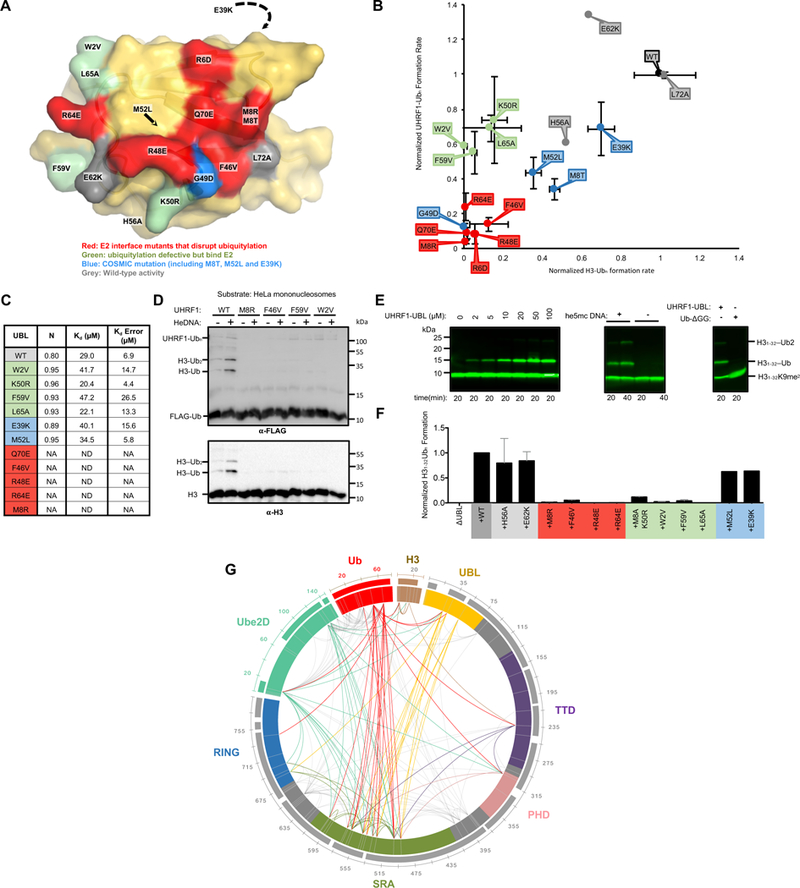
(A) Surface representation of the UBL domain with the mutation sites colored as indicated.
(B) Rates of ubiquitylation (see Methods) for assays containing the indicated mutants of UHRF1. Ubiquitylated substrate (H31–32K9me2; H31–32) and auto-ubiquitylated E3 products are both visualized in this dot plot. Errors reflect the standard deviation from biological duplicates; representative gels from which these rates were determined are in Supplemental Figure 5A.
(C) Kd values for indicated UBL variants binding to Ube2D1 as determined by ITC. Isotherms shown in Supplemental Figure 5B.
(D) HeLa mononucleosome ubiquitylation assay using WT or UHRF1 mutants (see Methods).
(E) H3 peptide (H31–32K9me2; H31–32) ubiquitylation assays in the presence of he5mc-DNA with ΔUBL and increasing UHRF1-UBL added in trans (left). The rescue of ΔUBL in the absence or presence of he5mc-DNA with 100 μM UHRF1-UBL (middle). (Right) ΔUBL rescue in the presence of 400 μM UHRF1-UBL or 400 μM ubiquitin (residues 1–74, Ub-ΔGG). Western blot is for biotin on the H3 peptide.
(F) Densitometry quantification of H3 substrate (H31–32K9me2-biotin) ubiquitylation ΔUBL rescue experiments with the indicated UHRF1-UBL mutants (see Supplemental Figure 6D).
(G) Chemical cross-linking/mass spectrometry analysis of the complex formed between UHRF1, he5mc-DNA, H3-peptide, and isopeptide-linked Ube2D-Ub conjugate. Crosslinks with q-values ≥ 0.01 are shown. Links that involve flexible linker regions of UHRF1 are in gray; those that involve structured domains are color-coded by domain (priority is given to the N-terminal-most domain). Sequence coverage of the MS data is shown as bars outside the circle plot.
