Figure 2.
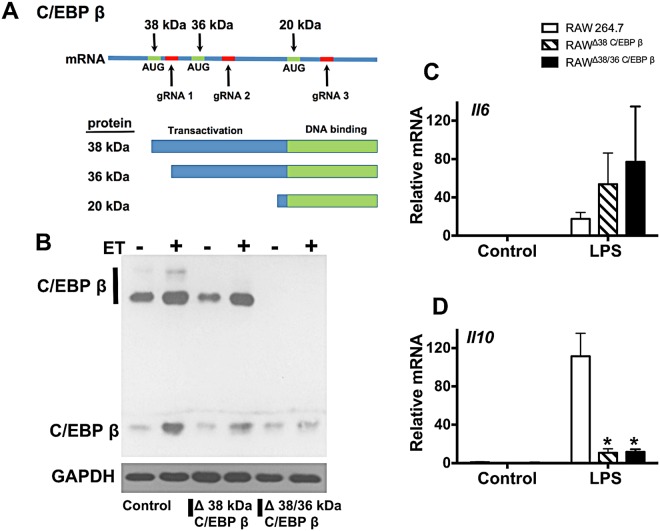
Isoform specific gene editing of C/EBP β modifies inflammatory responses in macrophages. Panel (A) Layout of the transcript that produces 3 separate protein isoforms termed 38 kDa C/EBP β, 36 kDa C/EBP β, and 20 kDa C/EBP β. The location of the 3 translation start sites (green) is depicted in relation to where the gRNAs target DNA for gene editing (red). Guide RNA 1 is used to produce a cell line lacking 38 kDa C/EBP β (RAWΔ38 C/EBP β). Guide RNA 2 is used to make a cell line lacking 38 and 36 kDa C/EBP β (RAWΔ38/36 C/EBP β). Guide RNA 3 produces a cell line lacking all 3 isoforms (RAWΔtotal C/EBPβ). Also presented is the domain layout of 3 C/EBP β protein isoforms showing differences in transactivation domain. Panel B–D. RAW 264.7 cells were transduced with lentivirus containing Cas9 and gRNA, and then clones were selected that do not produce 38 kDa C/EBP β (RAWΔ38 C/EBP β) or 38/36 kDa C/EBP β (RAWΔ38/36 C/EBP β). Panel (B) Demonstration of changes to C/EBP β isoform expression as shown by immunoblot analysis of C/EBP β and GAPDH. To increase expression of C/EBP β, these cell lines were exposed to ET (10 nM of EF and 10 nM of PA) for 6 h. Panel (C,D) RAWΔ38 C/EBP β cells, RAWΔ38/36 C/EBP β cells, and control RAW 264.7 cells were exposed to 1 μg/ml LPS for 6 h. RT-qPCR was used to quantify transcript levels of Il6 and Il10. RT-qPCR data are presented as mean (n = 3) ± S.D. Asterisks indicate significant different than control cells. *p < 0.05.