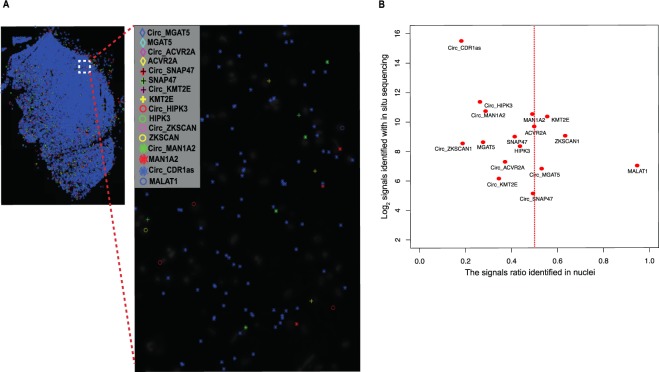
Figure 6.
Multiplex detection of circRNAs and linear RNAs with padlock probes in combination with in situ sequencing in human brain tissue. (A) Padlock probes, RCA, and in situ sequencing were performed to detect the cellular localization of circRNAs and linear RNA in a tissue section from fresh/frozen human brain (sample 5). CircRNAs and their corresponding linear RNA from 7 genes were screened (MGAT5, ACVR2A, SNAP47, KMT2E, HIPK3, ZKSCAN, and MAN1A2). In addition, circRNA from CDR1as and linear RNA from MALAT1 (known to be exclusively expressed in the nucleus, and used as a control for the localization experiment) were also included. Padlock probe sequences for each of the targets are listed in Supplementary Materials. In situ sequencing was performed to identify the 4-bp specific sequence barcodes in RCA products (RCPs) of each of the targets as shown in43. All targets were plotted on the DAPI image of the brain tissue section after in situ sequencing. Localization of each of the detected targets is shown as a symbol, left panel. Each symbol represents a barcode sequence that corresponds to a specific target. The nuclei are shown in grey. Scale barrepresents 100 μm. Zoom in region corresponds to the region in white dash-line square. (B) Abundance and subcellular localization of circRNA and linear RNAs. The ratios of signals in nuclei for all targets were plotted against their barcode counts in the brain sample. Similar to the circRNA panel data, CDR1as was the most abundant circRNAs in the in situ sequencing experiment. Also, in agreement with previous data, MALAT1 was highly expressed in the nucleus in the in situ sequencing experiment.