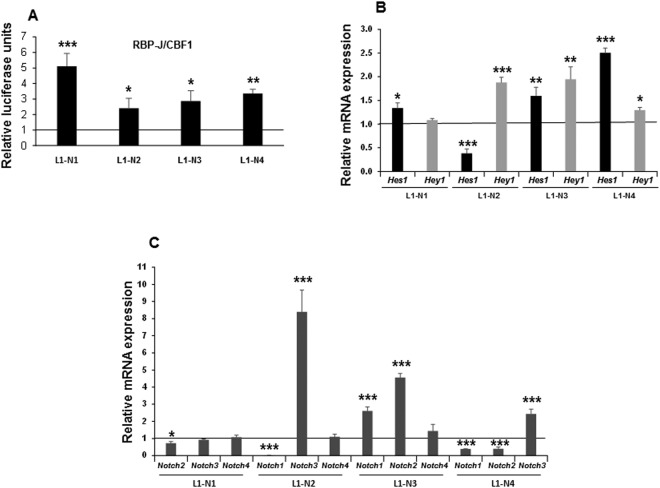
Figure 7.
NOTCH activation and signaling in 3T3-L1 cells stably over-expressing each one of the four NOTCH receptors. (A) qRT-PCR analysis of the relative Hes1 and Hey1 mRNA expression levels in the stable Notch1 gene transfectant (L1-N1), the stable Notch2 gene transfectant (L1-N2), the stable Notch3 gene transfectant (L1-N3), and the stable Notch4 gene transfectant (L1-N4). (B) NOTCH transcriptional activity, as measured by gene reporter luciferase assays, in these four Notch genes stable transfectants. (C) qRT-PCR analysis of the relative individual Notch mRNA expression levels in stable Notch1 gene transfectant (L1-N1), the stable Notch2 gene transfectant (L1-N2), the stable Notch3 gene transfectant (L1-N3), and the stable Notch4 gene transfectant (L1-N4). The relative luciferase activities were always normalized with renilla values and referred to those of control cells. Data in all qRT-PCR assays were normalized to P0 mRNA expression levels. The fold activation or inhibition in all assays is measured relative to the empty vector control, set arbitrarily at 1. Data are shown as the mean ± SD of at least three biological assays performed in triplicate. The statistical significance of Student’s t-tests results is indicated (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).