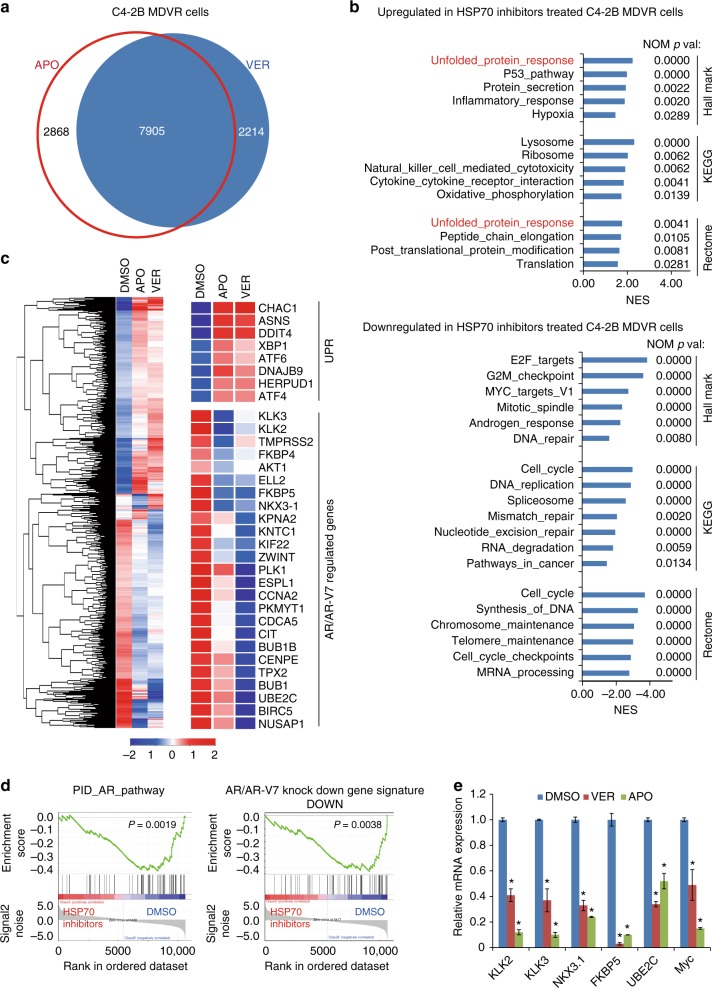
Fig. 5.
HSP70 inhibitors suppress AR and AR-V7 signaling. a Venn diagram of RNA-seq analysis of the two comparisons: APO vs. DMSO and VER vs. DMSO in C4-2B MDVR cells. b GSEA of top enriched gene sets in C4-2B MDVR cells treated by HSP70 inhibitors. The upregulated and down regulated gene sets from the Hallmark, KEGG and Rectome platforms were output by GSEA. c Heatmap and hierarchical clustering of the differentially expressed genes (DEGs) between APO and VER treatment in C4-2B MDVR cells with fold change (FC) > 1.2, as compared to vehicle (DMSO). The genes were displayed in rows and the normalized counts per sample were displayed in columns. Red indicates up-regulated and blue designates down-regulated expression levels. Middle and right, UPR, AR, and AR-V7 activity-signature genes that were altered in expression are displayed. d GSEA of the PID-AR pathway in C4-2B MDVR cells treated with HSP70 inhibitors, as compared to DMSO (left). GSEA of the AR and AR-V7 gene signatures in C4-2B MDVR cells treated with HSP70 inhibitors (right). The signature was defined by genes that underwent significant expression changes as a result of AR and AR-V7 knockdown in prostate cancer cells 6. e qRT-PCR analysis of the indicated genes in C4-2B MDVR cells treated with DMSO or with HSP70 inhibitors (10 μM) for 48 h. *p < 0.05. Results are the mean of three independent experiments (±s.d.). Statistical analysis was performed using two tailed Student’s t-test. APO apoptozole, VER Ver155008