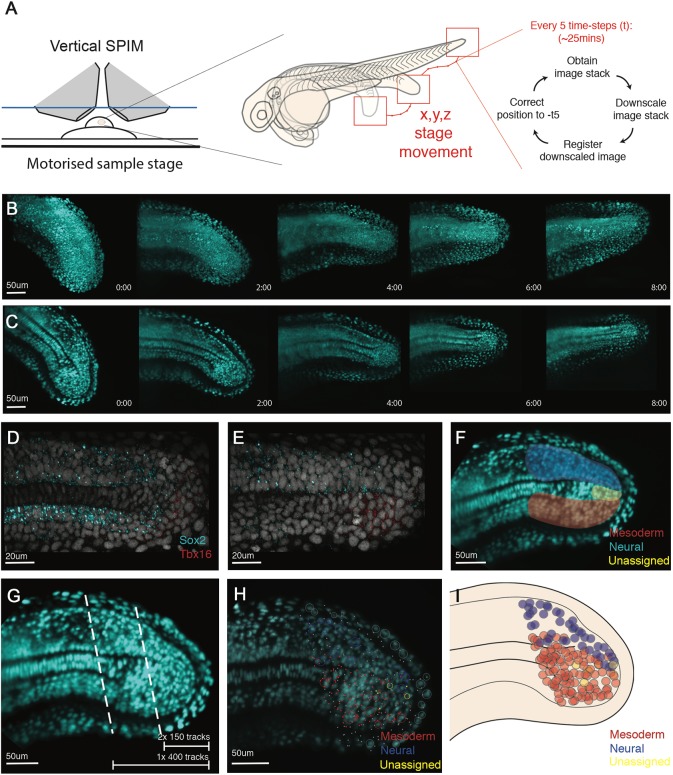
Fig. 7.
Single cell tracking of tailbud progenitors during late somitogenesis demonstrates an absence of bi-fated cells and cellular mixing within the tailbud. (A) Experimental design to allow long-term light-sheet imaging of the growing zebrafish tailbud. Embryos at the 21-somite stage were mounted as described by Hirsinger and Steventon (2017). Z-stacks were captured every 2.5 min with image-based registration on downsampled images conducted every five frames. The shift of this registration is then fed back into the stage position in all three directions to re-centre the tailbud within the image. (B,C) Individual frames are shown every 2 h across the 8 h movie shown as both maximum projections (B) and as a medial slice (C). (D-G) Fates of cells were assigned by the termination point of the track by scoring-based position within the anatomy of the tailbud and the associated expression of neural markers (Sox2) and mesdoermal markers (Tbx16 and Tbxta). Medial slices of hybridization chain reaction are shown in D and E, and used to zone the tailbud in F. Automatic tracks were selected using a custom MATLAB script by selecting all cells posteriorly to the lines shown in G. Two movies were subsetted using the most-posterior lines generating two movies of 150 tracks each. A third movie was subset using the more-anterior line, generating 400 tracks. (H,I) Final fates were assigned and have been overlaid over the starting timepoint (H) with open circles representing cells close to the viewed plane and dots representing cells on a different z-section. These different z-planes have been collapsed into a single 2D image (I) and demonstrate an absence of a mixed population of unfated cells. The mixing around the notochord is an artifact of collapsing the z-axis into a 2D image. No cells were observed dividing with progeny entering both lineages.