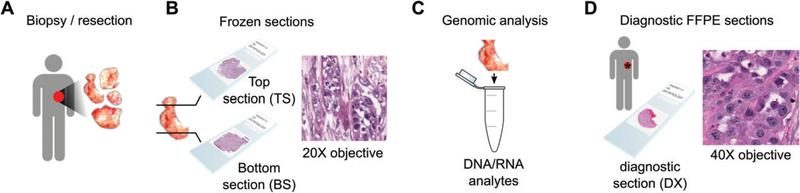
Figure 2.
Tissue procurement in TCGA. (A) A Tissue Source Site (TSS) obtains samples from surgical resection. (B) A portion of this tissue is selected for submission to TCGA, and the BCR produces ‘top-section’ (TS) and ‘bottom-section’ (BS) slides for review to determine that the percentage necrosis and abundance and proportion of tumour cells are adequate for genomic analysis. (C) The middle portion of this tissue is used to extract RNA and DNA analytes for genomic analysis. (D) One or more ‘diagnostic’ formalin-fixed paraffin-embedded (FFPE) slides are submitted to the BCR by the TSS for confirmation of histological diagnosis. These diagnostic slides originate from the same tumour, but their relationship to the material submitted for genomic analysis is unknown. The frozen sections provide the best representation of the tissue contents reflected in genomic signatures. However, the freezing artefacts in these slides can confound routine pathological examination or image analysis algorithms. The FFPE sections reveal cytological details, and have sufficient quality to confirm diagnosis, but the relationship or molecular similarity of these sections to the tissues submitted for genomic analysis is not as precise, as larger tumours may have considerable heterogeneity, and it is not always clear where the frozen tissue was sampled from relative to these H&E sections. The tradeoff between image quality and adjacency to genomic materials is an important consideration in designing an image analysis study of TCGA, and should be weighed on the basis of intratumoural heterogeneity and sensitivity of the image analysis algorithms to artefacts