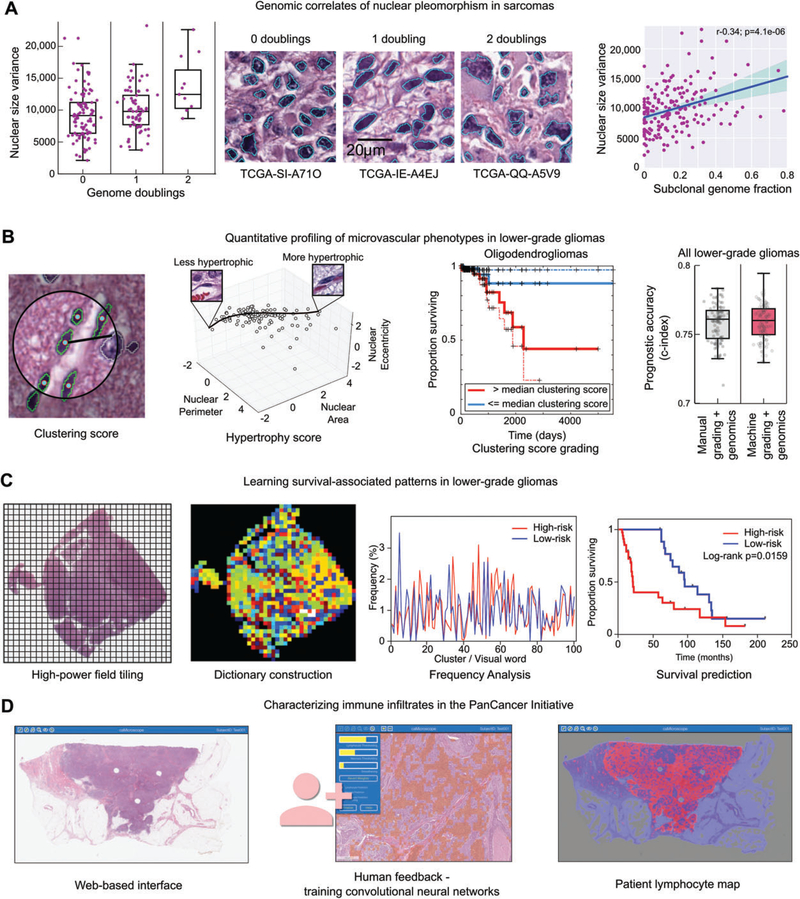
Figure 4.
Image analysis studies of TCGA. (A) Nuclear morphometry was used to study the genomic correlates of nuclear pleomorphism in sarcomas. Image segmentation was used to delineate >500 million nuclei in diagnostic sarcoma images, and the area of each nucleus was calculated. The variance of nuclear area was calculated for 235 sarcomas, and compared with measurements of genome doublings and subclonality obtained from sequencing and copy number data. Increased pleomorphism was significantly associated with measures of genomic complexity, including genome doublings, subclonality, and aneuploidy. (B) Machine learning was used to investigate microvascular phenotypes in lower-grade gliomas. A classifier was developed to identify vascular endothelial cells in gliomas (green). These classifications were used to measure to the clustering of endothelial cells and to model the morphological spectrum of endothelial nuclei in order to describe the extent of endothelial hyperplasia and hypertrophy in TCGA samples. These measurements were used as a biomarker to stratify overall survival, and were as effective at predicting outcomes as manual histological grading when combined with diagnostic genetic biomarkers. (C) Unsupervised machine learning was used to identify survival-associated patterns in lower-grade gliomas using TCGA data. Features describing the texture of haematoxylin were analysed in tiled high-power fields. These features were used to cluster the fields to define a dictionary of ‘visual words’ that captures the frequent patterns in the tissue. The frequency of these words in each slide were used to predict patient survival and to identify molecular correlates of histological patterns. (D) Convolutional networks were used to map the spatial distribution of TILs in 13 cancer types as part of the recent PanCancer immune working group. A web-based interface was used to train convolutional neural networks to identify patches containing TILs. These algorithms were then used to map the presence of TILs in >6000 whole slide images.