Figure 1. Six CRC subtyping systems derived from a single-omic classification workflow and two major strategies for integrative analysis. (a).
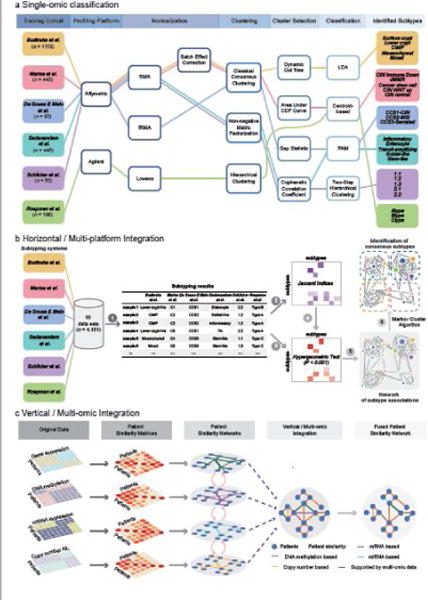
The workflow for six independent CRC subtyping systems based on single-omic classification strategy. The six CRC subtyping studies employed different training cohorts, gene expression profiling platforms, clustering and classification methods, yielding discrepant subtyping results. (b) We proposed a network-based approach for multi-platform (horizontal) integration, involving several major steps: (1) classifying 18 data sets totalling over 4000 samples using each of the six subtyping systems; (2) calculating a matrix of Jaccard indices quantifying the association between each pair of subtypes; (3) evaluating the statistical significance of the association between each pair of subtypes using hypergeometric tests; (4) filtering Jaccard indices by the p-values derived from hypergeometric tests to retain only significant associations (P < 0.001); (5) constructing a network of subtype associations; (6) partitioning the network into consensus molecular subgroups using Markov cluster algorithm (MCL) [82]. More technical details can be found in [9]. (c) Multi-omic (vertical) integration for cancer classification using similarity network fusion (SNF) [80]. Two or more types of omic data such as mRNA expression, miRNA expression, DNA methylation and copy number profiles can be integrated for more comprehensive dissection of cancer heterogeneity occurring at multiple omic levels of gene regulations.
