Abstract
The rapid drop in the cost of DNA sequencing led to the availability of multi-gene panels, which test 25 or more cancer susceptibility genes for a low cost. Clinicians and genetic counselors need a tool to interpret results, understand risk of various cancers, and advise on a management strategy. This is challenging as there are multiple studies regarding each gene, and it is not possible for clinicians and genetic counselors to be aware of all publications, nor to appreciate the relative accuracy and importance of each. Through an extensive literature review, we have identified reliable studies and derived estimates of absolute risk. We have also developed a systematic mechanism and informatics tools for (1) data curation, (2) the evaluation of quality of studies, and (3) the statistical analysis necessary to obtain risk. We produced the risk prediction clinical decision support tool ASK2ME (All Syndromes Known to Man Evaluator). It provides absolute cancer risk predictions for various hereditary cancer susceptibility genes. These predictions are specific to patients’ gene carrier status, age, and history of relevant prophylactic surgery. By allowing clinicians to enter patient information and receive patient-specific cancer risks, this tool aims to have a significant impact on the quality of precision cancer prevention and disease management activities relying on panel testing. It is important to note that this tool is dynamic and constantly being updated, and currently, some of its limitations include (1) for many gene-cancer associations risk estimates are based on one study rather than meta-analysis,(2) strong assumptions on prior cancers, (3) lack of uncertainty measures, and (4) risk estimates for a growing set of gene-cancer associations which are not always variant specific. All of these concerns are being addressed on an ongoing basis, aiming to make the tool even more accurate.
Keywords: Germline mutation, Disease susceptibility, Genetic predisposition to disease, Risk assessment, Risk management
Introduction
Until recently, patients with hereditary cancer susceptibility predisposition were tested for only a few well-known and studied genes, usually related to a single syndrome. For example, women at high risk of breast or ovarian cancer were tested only for BRCA1 and BRCA2. The rapid drop in the cost of DNA sequencing, as well as the over-turning of patents, have led to the availability of multi-gene cancer panels, which test a large number of genes at a relatively low cost (Plichta et al. 2016). These panels can test 25 or more cancer susceptibility genes at a time, often at a lower cost than previous single syndrome testing.
Following testing, patients rely on providers to interpret results, assess risk of various cancers, and advise on a management strategy. Evidence on the types of cancer associated with these genes, and the magnitude of the risk, is emerging rapidly (Couch et al. 2014; Desmond et al. 2015; Maxwell et al. 2014; Plichta et al. 2016; Tung et al. 2015, 2016a, 2016b; Walsh et al. 2011). However, the number of gene-cancer associations is large, information is dispersed over a vast number of studies, the quality of the studies is uneven, and the data presented are seldom directly applicable to patient care (e.g., one may find hazard ratios instead of the necessary absolute risk estimates). In an environment in which there is constant pressure to increase efficiency and see more patients, it is very challenging for clinicians to be aware of all publications, nor to appreciate the relative accuracy and importance of each. In addition, new information is becoming available on almost a daily basis, often revising prior beliefs. Aside from a few cases, such as BRCA1 and BRCA2 or the MMR genes, clinicians lack simple and reliable tools to personalize risk prediction, or to help make prevention decisions, for individuals who are found to carry deleterious mutations.
At present, laboratory genetic testing reports include generalized recommendations for management for patients with that particular mutation; however, these recommendations do not necessarily apply to the individual patient. For example, a patient with a PALB2 mutation, at young age, has about a 45% lifetime risk of developing breast cancer. But that is not true if she is 70 years old, or if she has had risk reducing mastectomies, or if she is 35 and has had her ovaries removed. Gene mutations will have a different significance based on the age of the patient, on the gender of the patient, on whether the patient has had organs removed in the past (e.g., hysterectomy or bilateral mastectomy), and on whether the patient has had cancer in the past (Ma et al. 2017; Mai et al. 2016; Riley et al. 2012). For some genes, it is becoming apparent that risk of disease also varies by the specific pathogenic variant of a mutation (Cybulski et al. 2007; Thompson et al. 2001). What a provider needs are risk predictions and management options for his or her specific patient.
Effective strategies for prevention or early detection of some cancers (for example, colorectal cancer) are available for high risk individuals, though few are feasible in an untargeted population-wide implementation. An important challenge is to use risk to personalize these prevention strategies. Accurate risk stratification could be used to guide the choice, frequency and age of onset of screening modalities and risk-reducing strategies. For this task, it is important to provide accurate risk stratification for individuals. While studies on gene-cancer associations are growing at a rapid rate, there is no central tool for clinicians to access data from these studies. To address this gap, we developed a systematic approach for (1) data curation, (2) quality assessment, (3) risk estimation, and (4) presentation of risk in an intuitive visualization specific to the patient. The result of this effort is a web-based clinical decision support tool, the All Syndromes Known to Man Evaluator ASK2ME.org.
Materials and Methods
Having identified the need for a tool which provides patient specific risk predictions for all cancer susceptibility genes, we identified the steps needed to create such a clinical decision support tool. These were as follows:
Development of a knowledge base structure and maintenance methodology
Literature review and data curation into the knowledge base
Quality assessment of individual studies
Risk estimation
Clinician facing interface
Development of a Knowledge Base Structure and Maintenance Methodology
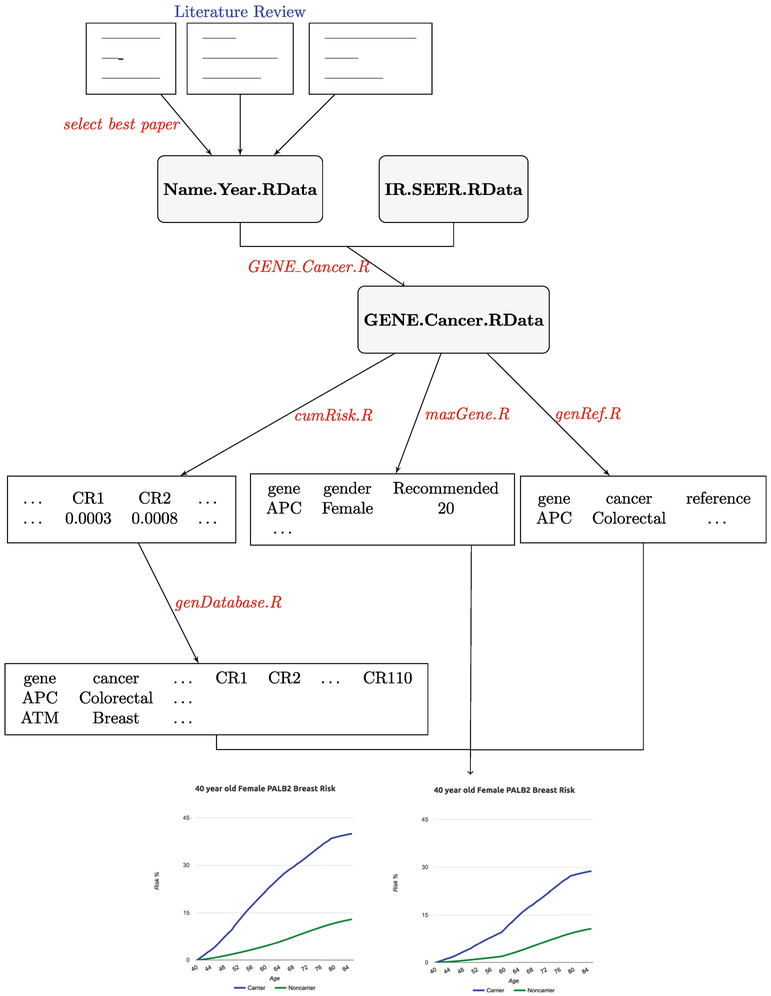
The first component of the infrastructure is the ask2meKnoweledgeBase package, written in the object-oriented and open source language R (R Core Team 2017). R is an environment for statistical computing and provides a large variety of statistical and graphical tools. The knowledge base was designed as a set of linked tables that store a machine-readable data set derived from the prose, tables, and figures of published literature regarding gene-cancer associations. We developed tools that facilitated many of the steps required to go from an article in its PDF format to this computable R object. Figure 1 provides an overview of these steps. Brief descriptions of the steps in Fig. 1 will be provided below as we introduce the infrastructure; detailed description of the abbreviations used in Fig. 1 can be found in Supporting Information, Appendix A. At the end of the process, each published study that was identified by search criteria and passed eligibility criteria was embedded in an R object with well-defined fields, which corresponds to Name.Year.RData in Fig. 1. These R objects contain prose, tables, or figures of the selected studies named by the first author’s last name and the year of publication.
Fig. 1.
Flowchart describing the steps currently used to populate the knowledge base behind the ASK2ME decision support tool
Literature Review and Data Curation into the Knowledge Base
We assembled the relevant studies for each gene-cancer association using well-established systematic review approaches to searching literature databases (Haidich 2010; Khan et al. 2003). We performed PubMed and Embase searches, starting with the following keywords in the title/abstract of the articles: (Gene OR Synonyms) AND (Germline OR Germ-line OR Inherited OR Genetic Predisposition to Disease[MeSH Major Topic]) AND (Penetrance OR Incidence) AND (Cancers of Interest). Here, “Gene” refers to the specific gene considered, for example APC. We performed this literature review for all known gene-cancer associations by reviewing associations reported by genetic testing companies as well as those reported by Plichta et al. (2016). We continue to systematically search the literature for studies reporting new associations, as well as update existing ones, so that the knowledge base is dynamic and is updated on an ongoing basis.
Quality Assessment of Each Study
The goal of the quality assessment was (1) to select a single, sufficiently reliable, study to be used for the risk estimation for a specific gene-cancer association or (2) to conclude that no study is of sufficient quality and detail to provide risk estimates. We developed a paper ranking system to assess the quality of published studies for specific gene-cancer associations (Table 1), and, based on this system, which is based on the number of carriers, selected the highest quality study for each gene-cancer association. Unless the selected study is itself a meta-analysis, a major limitation of this approach is that we do not currently integrate information across published studies (this is a future goal, see “Discussion”). We required the highest quality study to have a ranking of three stars or higher. If more than one study had a similar ranking, we chose the study with the larger sampler size. We defined lack of sufficient evidence as being unable to identify studies with a ranking of three stars or higher. In addition, we developed a gene-cancer association ranking (Table 1) to assess the strength of association. This ranking is based on both the number of carriers in the final paper as well as the number of studies on the gene-cancer association in the literature. After studies were ranked based on their quality, appropriate studies were presented and discussed at our combined lab meeting to confirm ratings and clinical utility. The best study was then chosen based on the final ranking, as well as study design and statistical considerations. Recently, Strande et al. (2017) developed a semi-quantitative metric based on ClinGen and the Developmental Disorder Genotype-Phenotype (DDG2P) database which also ranks gene-cancer associations, but differs from our ranking which is based on literature review and the number of carriers in the final paper.
Table 1.
Literature ranking system used to rank studies and associations
| Rating | Paper ranking |
|---|---|
| 1 | Number of carriers < 50 |
| 2 | Number of carriers is ≥ 50 and < 100 |
| 3 | Number of carriers is ≥ 100 and < 200 |
| 4 | Number of carriers is ≥ 200 and not a meta-analysis |
| 5 | Robust meta-analysis |
| Gene cancer association ranking | |
| 1 | Number of carriers in final paper ≤ 50 and number of gene/cancer association studies ≤ 3 |
| 2 | Number of carriers in final paper ≤ 50 and number of gene/cancer association studies > 3 |
| 3 | Number of carriers in final paper > 50 and number of gene/cancer association studies ≤ 3 |
| 4 | Number of carriers in final paper > 50 and number of gene/cancer association studies > 3 |
| 5 | Meta-analysis |
Highest quality study is one with a paper ranking of three or higher. Studies with paper ranking of one or two provide insufficient evidence for risk estimation
Risk Estimation
After storing data from the best available study for each gene-cancer association in the ask2meKnowledgeBase R package, we then estimate the penetrance (e.g., the probability that an individual will develop the disease by a specific age), for each gene-cancer association by gender.
Most studies do not report the penetrance risk estimates directly, but instead report other measures of risk including odd ratios (ORs), hazard ratios (HRs), relative risks (RRs), standardized incidence ratios (SIRs), and cumulative risk (CR) estimates. We developed statistical methodology to estimate the penetrance from these measures, described in detail in Supporting Information, Appendix B. For studies that do report penetrance estimates directly (cause-specific penetrance, cause-specific survival, or cause-specific cumulative incidence), we are able to use these quantities directly.
For studies reporting one of the following measures: OR, HR, RR, and SIR comparing gene mutation carriers to non-carriers, we assume that the reported ratio is constant throughout the patients’ lifetime (unless the study report these quantities stratified by age). In order to estimate penetrance for carriers, we then combine the reported ratio with the baseline risk for noncarriers for each cancer. We obtain this baseline risk estimates from SEER (Surveillance, Epidemiology, and End Results Program), which provides the age-specific probability for each cancer by gender through their DevCan software. We obtain data from SEER 18 Registries, which we input into their DevCan software and store the output as an R object, IR.SEER.RData (Fig. 1).
We incorporated a selected subset of prior cancers and prior surgeries into the risk estimation and into the database. For now, we only adjust for prior cancer by removing the selected cancer when displaying the future risk. This is a very strong assumption, as we assume that (1) prior cancer does not impact the risk of developing other cancers, and (2) prior cancer does not impact the risk of recurrence or second primary cancer (this assumption is discussed further in the “Discussion”). For prior surgeries, we have incorporated mastectomy, oophorectomy, and hysterectomy, all with strong assumptions, that require further extensions (Table 2). We assume hysterectomy reduces the risk of endometrial cancer for carriers and noncarriers of any gene to 0 but has no impact on the risk of other cancers. For oophorectomy and mastectomy, we use established estimates of risk reduction for BRCA1/2 carriers and noncarriers for breast and ovarian cancers (Katki 2007), and assume that noncarriers and carriers of other genes who have these surgeries have the same risk reduction as the previously estimated reduction for noncarriers.
Table 2.
Risk reduction multiplier for mastectomy, oophorectomy, and hysterectomy, by gender and/or carrier status
| Prior surgery | Carrier status | |||||
|---|---|---|---|---|---|---|
| Mastectomy | Oophorectomy | Hysterectomy | BRCA1 carriers | BRCA2 carriers | Noncarriers and other gene mutation carriers | |
| Female | ||||||
| Breast cancer | Yes | Yes | – | 0.0511 | 0.0361 | 0.0461 |
| Yes | No | – | 0.12 | 0.12 | 0.12 | |
| No | Yes | – | 0.513,a | 0.363,a | 0.46a,4 | |
| Ovarian cancer | – | Yes | – | 0.313 | 0.283 | 0.054 |
| Endometrial cancer | – | – | Yes | 05 | 05 | 05 |
| Male | ||||||
| Breast cancer | Yes | – | – | 0.15 | 0.15 | 0.15 |
By multiplier, we mean that the risk estimates are multiplied by the factors in this table based on mutation carrier status and the prophylactic surgeries listed in this table
For risk modification for mastectomy for males and hysterectomy for females, risk is modified regardless of carrier status
There is not enough information from published papers for these combinations of prior surgeries. Current estimates are obtained by assuming that the joint effect of prophylactic surgeries can be estimated by the product of the effects of individual surgeries
We are using the risk reduction rates from conclusions in Rebbeck et al. (2004)
We are using the hazard ratios from Domchek et al. (2010). Ratios for oophorectomy and breast cancer are from Table 3; ratios for oophorectomy and ovarian cancer are from Table 2 in the paper
We are using the hazard ratios from “Incorporate Oophorectomy into BRCAPRO” section in Katki (2007)
There is not enough information from published papers for these combinations of prior surgeries. Current estimates are obtained using clinical subject matter knowledge
The protective effect of oophorectomy only applies if the patient is under the age of 45
To fully document the data extraction process, facilitate later modifications, and ensure reproducibility across potentially evolving team membership, we developed R functions specific for each study for a specific gene-cancer association, GENE_Cancer.R in Fig. 1. These functions will take in measures of risks reported in the studies stored in Name.Year.RData (described earlier), combine these risks with IR.SEER.RData if necessary, and finally output the penetrance estimates, stored as GENE.Cancer.RData in Fig. 1 (described in detail in the Supporting Information, Appendix C).
Clinician Facing Interface
We created a clinician facing interface (website), which allows users to select gene, gender, age, prior surgery, and prior cancer. After entering this information, the clinician is then provided with patient-specific risk estimates in figures and tables. In addition, next to each figure, we display the citation of the study from which the risk estimates were obtained. For some known gene-cancer associations, there is insufficient data to quantitatively estimate the risk, and these are listed in prose on the output as well.
We developed functions using GENE.Cancer.RData stored in the ask2meKnowledgeBase R package (Fig. 1) to create a database, which is the back end of the clinician interface website. genDatabase.R is created to store the risk estimates for all the gene-cancer associations given on the website, which utilizes cumRisk.R function to convert the age-specific risk estimates to age-conditional probabilities of developing cancer. maxGene.R is created to provide gene-gender level information displayed on the website, e.g., range for the cancer risk estimates for the same gene and gender, and lack of sufficient evidence for some cancers for a specific gene. genRef.R is created to provide gene-gender-cancer level information displayed on the website, e.g., references for each gene-cancer association and the corresponding ratings. More details are provided in the Supporting Information, Appendix C.
Results
The ASK2ME website provides patient specific risk estimates by gene-cancer gender for 65 gene-cancer associations (Tables 3 and 4). Please note that the tool is being updated on a monthly basis (details on the exact dates of the updates can be found in the updates log on the ASK2ME website), with these tables changing as the tool is updated. The risk for each gene-cancer association is based on extensive literature review identifying reliable studies on the cancer risk implication. For each association, studies are ranked, and risk estimates are based on the highest ranked study (Table 3). For some known associations, we were unable to identify reliable studies for risk estimation, that is we are unable to identify studies with a ranking of three stars or higher based on the ranking system developed (Table 1). These associations are still listed on the website with references.
Table 3.
References for most reliable literature for associated cancers by gene. Risk estimates in ASK2ME are provided based on the most reliable study for each association
Table 4.
Gene cancer associations currently included in ASK2ME
| Genes | Reliable studies for risk estimation | |
|---|---|---|
| Yes | No | |
| ATM | Breast (F), colorectal (F/M), gastric (F/M), pancreatic (F/M), prostate | Breast (M), leukemia (F/M), lymphoma (F/M) |
| BMPR1A | – | Colorectal (F/M), gastric (F/M), pancreatic (F/M), small bowel (F/M) |
| BRCA1 | Breast (F/M), ovarian, pancreatic (F/M) | Fallopian tube, peritoneal, prostate |
| BRCA2 | Breast (F/M), ovarian, pancreatic (F/M), prostate | Fallopian tube, peritoneal, melanoma (F/M) |
| BRIP1 | Ovarian | Breast (F) |
| CDH1 | Breast (F), gastric (F/M) | Colorectal (F/M) |
| CDK4 | Melanoma (F/M) | Pancreatic (F/M) |
| CDKN2A/p16 | Melanoma (F/M), pancreatic (F/M) | Melanoma (F/M), pancreatic (F/M) |
| CDKN2A/p14 | – | Melanoma (F/M), pancreatic (F/M) |
| CHEK2/1100delC | Breast (F), colorectal (F/M), prostate | Thyroid (F/M), ovarian, kidney (F/M) |
| GREM1 | – | Colorectal (F/M) |
| MLH1 | Colorectal (F/M), endometrial, gastric (F/M), ovarian | CNS (F/M), hepatobiliary (F/M), kidney (F/M), pancreatic (F/M), prostate |
| Sebaceous gland (F/M), skin (F/M), small bowel (F/M), ureter (F/M), urinary bladder (F/M) | ||
| MSH2 | Colorectal (F/M), endometrial, gastric(F), ovarian, pancreatic (F/M) | CNS (F/M), gastric (M), hepatobiliary (F/M), kidney (F/M), pancreatic (F/M), prostate |
| Sebaceous gland (F/M), small bowel (F/M), ureter (F/M), urinary bladder (F/M) | ||
| MSH6 | Colorectal (F/M), endometrial, gastric (F/M), ovarian | CNS (F/M), hepatobiliary (F/M), kidney (F/M), pancreatic (F/M), prostate |
| Sebaceous gland (F/M), small bowel (F/M), ureter (F/M), urinary bladder (F/M) | ||
| MUTYH-Biallelic/G396D | Colorectal (F/M) | – |
| MUTYH-Biallelic/CH | Colorectal (F/M) | – |
| MUTYH-Biallelic | Ovarian, urinary bladder (F/M) | Breast (F), small bowel (F/M) |
| MUTYH-Monoallelic | Colorectal (F/M), endometrial, gastric (F/M), hepatobiliary (F/M) | Breast (F), small bowel (F/M) |
| NBN/657del5 | Breast (F), prostate | Ovarian, skin (F/M) |
| PALB2 | Breast (F), prostate, pancreatic (F/M) | Breast (M) |
| PMS2 | Colorectal (F/M), endometrial, ovarian | CNS (F/M), gastric (M), hepatobiliary (F/M), kidney (F/M), pancreatic (F/M), prostate |
| Sebaceous gland (F/M), small bowel (F/M), ureter (F/M), urinary bladder (F/M) | ||
| POLD1 | – | Colorectal (F/M) |
| POLE | – | Colorectal (F/M) |
| PTEN | Breast (F), colorectal (F/M), endometrial, kidney (F/M), melanoma (F/M), thyroid (F/M) | Urinary bladder (F/M) |
| RAD51C | Ovarian | Breast (F) |
| RAD51D | Ovarian | Breast (F) |
| SMAD4 | – | Colorectal (F/M), gastric (F/M), pancreatic (F/M), small bowel (F/M) |
| STK11 | Breast (F), colorectal (F/M), gastric (F/M), pancreatic (F/M) | Lung (F/M), endometrial, ovarian, small bowel (F/M) |
| TP53 | Breast (F), brain (F/M), leukemia (F/M), osteosarcoma (F/M), soft tissue (F/M) | Lung (F/M), endometrial, gastric (F/M), melanoma (F/M), ovarian, prostate |
Penetrance estimates are provided for associations for which quality of studies was high enough to estimate risk. A list of cancer associated with a specific gene, for which there is not enough evidence to estimate risk is also provided
CNS central nervous system, MUTYH-Biallelic/CHMUTYH-biallelic/compound heterozygous
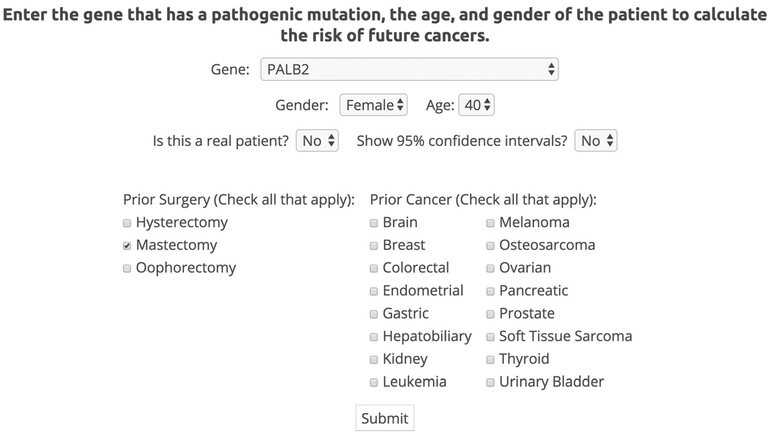
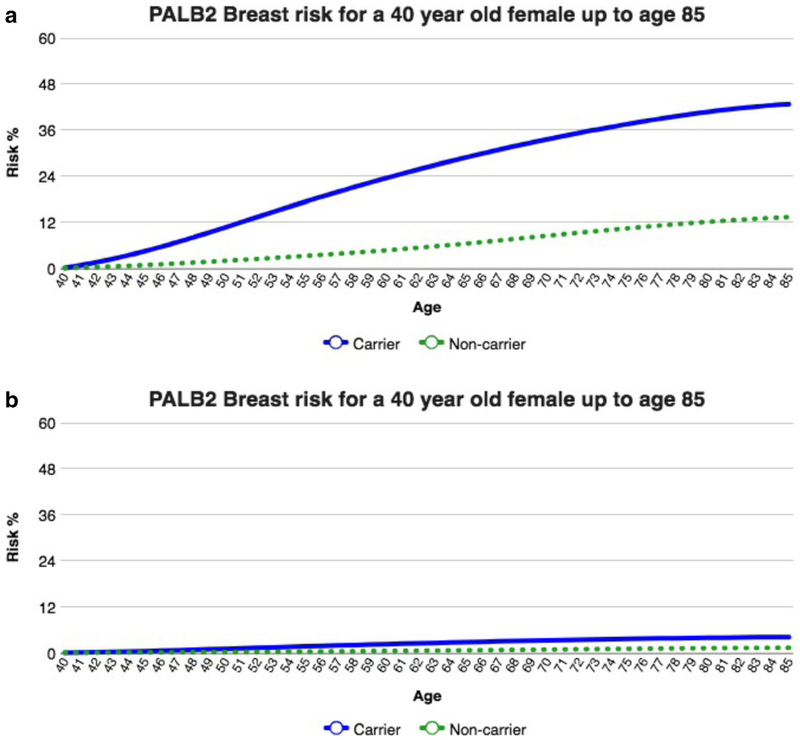
The website allows users to enter patient-specific characteristics including age, gender, prior cancers, prior surgeries, and gene of interest (Fig. 2). Based on these patient-specific characteristics, the website currently provides patient specific risk estimates in graphs and tables over age, starting at the patient’s current age (Fig. 3). Clinicians can look at the patient’s risk of cancer at each age for carriers and noncarriers of a specific mutation. For each gene-cancer association, we estimate the penetrance curve for the general carriers of a specific genetic mutation and for the general population. Two penetrance curves are illustrated in Fig. 3, highlighting the implication of prophylactic surgery (in this case mastectomy) on future risk.
Fig. 2.
ASK2ME user input. The user enters patient information on gene, gender, age, prior surgeries, and prior cancers
Fig. 3.
Risk of breast cancer for a 40-year-old female carrier of a deleterious germline mutation of the PALB2 gene, a without mastectomy and b with mastectomy
In clinical terms, these curves provide absolute risk estimates that are the foundation of decision support. The tool we developed will estimate the penetrance for each of the gene-cancer associations, and tailor it to an individual’s age and clinical history. Please note that although not displayed in Fig. 3, our tool displays below each figure the reference and ranking of the study which was used to estimate the figure, allowing clinicians to access those original papers easily.
Discussion
We developed the ASK2ME methodology and decision support tool with two goals in mind: (1) to provide an urgently needed systematization of the information used to guide decision making after panel testing; (2) to lay the foundation for scaling this process as both the number of genes and the relevant papers grows.
Accordingly, the ASK2ME website will require continuing updates, and we have designed the infrastructure of the knowledge base to allow for maintenance, enhancements, and extensions. As part of the ongoing work, we are also developing natural language processing algorithms to automate some of the updates and maintenance of the tool. In terms of extensions, the first extension is to other gene-cancer associations. To date, we have covered 29 genes, but other gene associations remain to be included. In addition, as new associations between cancers and genes are discovered, we will undertake a similar literature review, curation, rating, and analysis as described above and update the knowledge base appropriately, as well as conduct updates to current gene-cancer associations. The tool has version control and an updates log, which allows users to keep track of these ongoing updates. In addition, our long-term maintenance plan includes systematic review of each gene-cancer association at least once a year, and the formation of an advisory board to handle contradictory conclusions and ensure that the reviews are complete and upto-date.
We have taken the relatively straightforward approach of assuming that for most genes all pathogenic variants in a given gene will have the same implications on risk. However, it has become apparent that different pathogenic variants of the same gene may have different disease spectra and different penetrance (Groden et al. 1991; Rebbeck et al. 2015). We plan to extend the website to manage variant level risk as well, though this prospect is very challenging. As the list of genes and cancers increases, so will the list of variants, which range from pathogenic or likely pathogenic to variants of unknown significance (VUSs). In order to handle this growing list of variants, we need to devise automated approaches to constantly scan the literature in order to capture these changes.
While we believe that choosing and displaying the results of a single study available of sufficient quality has been a reasonable starting point, we realize that combining multiple studies via a meta-analysis has significant advantages. To this end, we are in the process of conducting a meta-analysis for each gene-cancer association when multiple quality studies are available. When estimating the penetrance using meta-analysis, in addition to taking into account each study’s design, there will be additional complexities. Since studies identified in the literature are unlikely to report penetrance directly, and instead report other measures of risk such as OR, HR, RR, SIR, and CR, conducting meta-analysis is not straightforward. Marabelli and co-authors (Marabelli et al. 2016) developed a likelihood-based approach to combine studies reporting penetrance, OR, RR, and SIR, which they apply to estimate the penetrance of breast cancer for ATM carriers. We are extending this approach to incorporate, in addition, studies reporting HR and CR.
Estimating the uncertainty of our risk estimates is crucial. One limitation of our tool is that the outputs displayed for clinicians on our website do not currently include confidence intervals for the estimates, though we are currently working on adding the confidence intervals to the plots, and this should be available soon. While we recognize the weaknesses of displaying risk estimates without confidence intervals, clinicians, nurses and genetic counselors are caring for patients with these pathogenic mutations now, and have few tools to assist them. We are hopeful that in the meantime, even without estimates of uncertainty, presenting a visualization of the best data available can still help clinicians make better decisions. For example, the level of risk for colon cancer with an APC mutation compared that of an MSH2 mutation can help explain why prophylactic surgery may useful for one but not necessary for the other, depending on patient’s preferences. While the visualizations we produce may also be used by patients directly, we want to emphasize that ASK2ME is designed for clinicians and not as a patient facing site. We can leverage the same knowledge base to produce a patient facing interface after sufficient design and usability analysis, but we have not done so to date.
A next natural step is to validate these risk estimates. Since many of these mutations are less common, a very large database of individuals would need to be collected in order to validate these risk estimates. We are not aware of any large-scale, publicly available cohorts that include panel testing results with the associated clinical information necessary for validation. As panel testing becomes more widely available, we hope that this work will motivate the creation and sharing of such databases.
Another limitation of the ASK2ME tool is that it does not yet estimate the risk of a second cancer of a given organ when a prior cancer has occurred. The website does not display future risk for that cancer and assumes that there is no impact on the risk of other cancers. Both of these assumptions may be unrealistic. The risk of second primary cancers can vary dramatically based on the initial treatment of the first cancer (bilateral mastectomy or the use of tamoxifen would both decrease the risk of second primary breast cancers). Also, the treatment of one cancer could potentially decrease the risk of cancers of other organs (ovarian cancer at a young age, treated by oophorectomy, will decrease breast cancer risk). Estimating separate penetrance curves conditional on prior cancer and its treatment will be crucial in estimating accurate risk for a patient (Davies et al. 2015; Trialists’ Group 2005).
Extending this tool to provide more accurate and patient-specific risk estimates by allowing users to enter additional characteristics, such as the race of the patient, whether or not the patient is of Ashkenazi Jewish descent, and the specific pathogenic variant is the subject of future work. Adding additional inputs will improve the output of the plots and tables and make the tool more usable in clinical practice, once sufficient data is available and curated.
Incorporating patient-specific management recommendations will enhance the usability of the tool, and we have begun working on this. Providing clinicians with summaries of the current guidelines is a crucial step for increasing the impact of screening and prevention approaches. Clinicians, especially in primary care, are often pressed for time and do not have the resources to keep abreast of current guidelines. The guidelines for clinicians are being updated on a regular basis often changing drastically from 1 year to another. Guidelines can even conflict from one source to another, as sometimes do guidelines from multiple government agencies, insurance companies, professional organizations, and researchers involved in writing guidelines. Our current strategy is to report guidelines from various sources, which currently include (1) National Comprehensive Cancer Network (NCCN), (2) European Society of Medical Oncology (ESMO) Clinical Practice Guidelines for Cancer Prevention and Screening, (3) publication by Tung et al. 2016a, 2016b, and (4) publication by Graffeo et al. (2016). Note, we do not attempt to assess the quality of these guidelines; rather, we curate the guidelines for clinicians.
Lastly, producing information sheets for patients, letters to referring clinicians, and formatted text to be incorporated in the EHR, will also increase the tool’s utility. Clinicians would be able to print these letters and give to them patients, to help guide patients in their decision-making. This feature has the potential of contributing to more uniform and effective communication of results across practitioners and allows patients to understand their results better, share results with family members, and improve disease prevention.
There is growing literature on risk communication and decision support tools. Sim et al. (2001) provide recommendations for clinical decision support systems, and this tool indeed follows some of the recommendations including capturing “literature-based and practice-based evidence in machine-interpretable knowledge bases” and developing “maintainable technical and methodological foundations for computer-based decision support.” Shared decision making is becoming a routine component of medical practice, and Hanoch et al. (2015) show that numeracy skills affect a patient’s desire to be involved in the decision making process. In addition, Portnoy et al. (2010) study the impact of numeracy skills and health literacy on the ability to learn information during a genetic counseling session and showed that both play an important role of learning information communicated during a session. The decision making process for prophylactic mastectomy among breast cancer patients with no BRCA mutations has been studied by Rendle et al. (2015). They identified various factors that influenced this decision making process, including subjective evaluations of risks and benefits. Risk communication is crucial, as numeracy skills of patients can vary dramatically. Studies such as Brewer et al. (2012) have explored the best format to communicate risk for breast cancer recurrence, and we plan to take such studies into account when designing a patient facing interface.
ASK2ME has the potential of having a direct clinical impact by providing a clinical decision support system that allows the clinician to enter the important factors about a patient with a pathogenic variant and immediately receive the risk of various cancers for that mutation for that patient with that current clinical situation. These risk predictions can in turn lead to more personalized prevention and management for individuals undergoing susceptibility testing and potentially help optimize screening and behavioral interventions to reduce risk. Future studies will include quantifying and evaluating the potential impact of the tool.
Supplementary Material
Acknowledgements
We gratefully acknowledge support from the National Cancer Institute at the National Institutes of Health [4P30CA006516–51] which supported Dr. Parmigiani.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10897-018-0238-4) contains supplementary material, which is available to authorized users.
Compliance with Ethical Standards
Conflict of Interest Dr. Hughes receives Honoraria from Myriad Genetics Veritas Genetics, Advisory Board for Beacon (An RFID Biopsy Marker), and is a founder of and has a financial interest in Hughes Risk Apps, LLC. Dr. Hughes’s interests were reviewed and are managed by Massachusetts General Hospital and Partners Health Care in accordance with their conflict of interest policies.
Dr. Parmigiani is a member of the Scientific Advisory Board and has a financial interest in Cancer Risk Apps LLC (CRA). CRA commercializes software for management of patients at high risk of cancer. At the present time, CRA is not supporting or licensing ask2me. We feel there is no significant overlap with this work.
Dr. Braun, Ms. Yang, and Ms. Griffin declare that they have no conflict of interest.
Human Studies and Informed Consent This article does not contain human subjects.
Animal Studies No animal studies were carried out by the authors for this article.
References
- Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, Lee A, Subramanian D, de Leeneer K, Fostira F, Tomiak E, Neuhausen SL, Teo ZL, Khan S, Aittomäki K, Moilanen JS, Turnbull C, Seal S, Mannermaa A, Kallioniemi A, Lindeman GJ, Buys SS, Andrulis IL, Radice P, Tondini C, Manoukian S, Toland AE, Miron P, Weitzel JN, Domchek SM, Poppe B, Claes KBM, Yannoukakos D, Concannon P, Bernstein JL, James PA, Easton DF, Goldgar DE, Hopper JL, Rahman N, Peterlongo P, Nevanlinna H, King MC, Couch FJ, Southey MC, Winqvist R, Foulkes WD, & Tischkowitz M (2014). Breast-cancer risk in families with mutations in PALB2. New England Journal of Medicine, 371(6), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow E, Robinson L, Alduaij W, Shenton A, Clancy T, Lalloo F, Hill J, & Evans DG (2009). Cumulative lifetime incidence of extracolonic cancers in lynch syndrome: A report of 121 families with proven mutations. Clinical Genetics, 75(2), 141–149. [DOI] [PubMed] [Google Scholar]
- Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, Guimbaud R, Buecher B, Bignon YJ, Caron O, Colas C, Noguès C, Lejeune-Dumoulin S, Olivier-Faivre L, Polycarpe-Osaer F, Nguyen TD, Desseigne F, Saurin JC, Berthet P, Leroux D, Duffour J, Manouvrier S, Frébourg T, Sobol H, Lasset C, Bonaïti-Pellié C, & French Cancer Genetics Network. (2011). Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA, 305(22), 2304–2310. [DOI] [PubMed] [Google Scholar]
- Brewer NT, Richman AR, DeFrank JT, Reyna VF, & Carey LA (2012). Improving communication of breast cancer recurrence risk. Breast Cancer Research and Treatment, 133(2), 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, & Parmigiani G (2007). Meta-analysis of BRCA1 and BRCA2 penetrance. Journal of Clinical Oncology, 25(11), 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, Olson JE, Godwin AK, Pankratz VS, Olswold C, Slettedahl S, Hallberg E, Guidugli L, Davila JI, Beckmann MW, Janni W, Rack B, Ekici AB, Slamon DJ, Konstantopoulou I, Fostira F, Vratimos A, Fountzilas G, Pelttari LM, Tapper WJ, Durcan L, Cross SS, Pilarski R, Shapiro CL, Klemp J, Yao S, Garber J, Cox A, Brauch H, Ambrosone C, Nevanlinna H, Yannoukakos D, Slager SL, Vachon CM, Eccles DM, & Fasching PA (2014). Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. Journal of Clinical Oncology, 33(4), 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Wokołorczyk D, Kładny J, Kurzwaski G, Suchy J, Grabowska E, Gronwald J, Huzarski T, Byrski T, Górski B, D bniak T, Narod SA, & Lubiński J (2007). Germline CHEK2 mutations and colorectal cancer risk: Different effects of a missense and truncating mutations? European Journal of Human Genetics, 15(2), 237–241. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Wokołorczyk D, Kluźniak W, Jakubowska A, Górski B, Gronwald J, et al. (2013). An inherited NBN mutation is associated with poor prognosis prostate cancer. British Journal of Cancer, 108(2), 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KR, Cantor SB, & Brewster AM (2015). Better contralateral breast cancer risk estimation and alternative options to contra-lateral prophylactic mastectomy. International Journal of Women’s Health, 7, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Snoo FA, Bishop DT, Bergman W, van Leeuwen I, van der Drift C, van Nieuwpoort FA, Out-Luiting CJ, Vasen HF, ter Huurne JAC, Frants RR, Willemze R, Breuning MH, & Gruis NA (2008). Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clinical Cancer Research, 14(21), 7151–7157. [DOI] [PubMed] [Google Scholar]
- Desmond A, Kurian AW, Gabree M, Mills MA, Anderson MJ, Kobayashi Y, Horick N, Yang S, Shannon KM, Tung N, Ford JM, Lincoln SE, & Ellisen LW (2015). Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncology, 1(7), 943–951. [DOI] [PubMed] [Google Scholar]
- Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, Garber JE, Neuhausen SL, Matloff E, Eeles R, Pichert G, van t’veer L, Tung N, Weitzel JN, Couch FJ, Rubinstein WS, Ganz PA, Daly MB, Olopade OI, Tomlinson G, Schildkraut J, Blum JL, & Rebbeck TR (2010). Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA, 304(9), 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowty JG, Win AK, Buchanan DD, Lindor NM, Macrae FA, Clendenning M, Antill YC, Thibodeau SN, Casey G, Gallinger S, Marchand LL, Newcomb PA, Haile RW, Young GP, James PA, Giles GG, Gunawardena SR, Leggett BA, Gattas M, Boussioutas A, Ahnen DJ, Baron JA, Parry S, Goldblatt J, Young JP, Hopper JL, & Jenkins MA (2013). Cancer risks for MLH1 and MSH2 mutation carriers. Human Mutation, 34(3), 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C, Loeffler M, Steinke V, Rahner N, Holinski-Feder E, Dietmaier W, Schackert HK, Goergens H, von Knebel Doeberitz M, Goecke TO, Schmiegel W, Buettner R, Moeslein G, Letteboer TGW, García EG, Hes FJ, Hoogerbrugge N, Menko FH, van Os TAM, Sijmons RH, Wagner A, Kluijt I, Propping P, & Vasen HFA (2012). Risks of less common cancers in proven mutation carriers with lynch syndrome. Journal of Clinical Oncology, 30(35), 4409–4415. [DOI] [PubMed] [Google Scholar]
- Graffeo R, Livraghi L, Pagani O, Goldhirsch A, Partridge AH, & Garber JE (2016). Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care. Breast Cancer Research and Treatment, 1–18. [DOI] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, Sargeant L, Krapcho K, Wolff E, Burt R, Hughes JP, Warrington J, McPherson J, Wasmuth J, le Paslier D, Abderrahim H, Cohen D, Leppert M, & White R (1991). Identification and characterization of the familial adenomatous polyposis coli gene. Cell, 66(3), 589–600. [DOI] [PubMed] [Google Scholar]
- Haidich AB (2010). Meta-analysis in medical research. Hippokratia,14(Suppl 1), 29–37. [PMC free article] [PubMed] [Google Scholar]
- Hanoch Y, Miron-Shatz T, Rolison JJ, Omer Z, & Ozanne E (2015). Shared decision making in patients at risk of cancer: The role of domain and numeracy. Health Expectations, 18(6), 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, Schrader KA, Schaeffer DF, Shumansky K, Zogopoulos G, Santos TA, Claro I, Carvalho J, Nielsen C, Padilla S, Lum A, Talhouk A, Baker-Lange K, Richardson S, Lewis I, Lindor NM, Pennell E, MacMillan A, Fernandez B, Keller G, Lynch H, Shah SP, Guilford P, Gallinger S, Corso G, Roviello F, Caldas C, Oliveira C, Pharoah PDP, & Huntsman DG (2015). Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncology, 1(1), 23–32. [DOI] [PubMed] [Google Scholar]
- Helgason H, Rafnar T, Olafsdottir HS, Jonasson JG, Sigurdsson A, Stacey SN, Jonasdottir A, Tryggvadottir L, Alexiusdottir K, Haraldsson A, le Roux L, Gudmundsson J, Johannsdottir H, Oddsson A, Gylfason A, Magnusson OT, Masson G, Jonsson T, Skuladottir H, Gudbjartsson DF, Thorsteinsdottir U, Sulem P, & Stefansson K (2015). Loss-of-function variants in ATM confer risk of gastric cancer. Nature Genetics, 47(8), 906–910. [DOI] [PubMed] [Google Scholar]
- Katki HA (2007). Incorporating medical interventions into carrier probability estimation for genetic counseling. BMC Medical Genetics, 8(1), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KS, Kunz R, Kleijnen J, & Antes G (2003). Five steps to conducting a systematic review. Journal of the Royal Society of Medicine, 96(3), 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang B, & Zheng W (2013). Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut, gutjnl-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma IT, Gray RJ, Wasif N, Butler KA, Cornella JL, Magrina JF, Magtibay PM, Casey WJ, Mahabir R, Rebecca AM, Hunt KS, & Pockaj BA (2017). Outcomes of concurrent breast and gynecologic risk reduction surgery. Annals of Surgical Oncology, 24(1), 77–83. [DOI] [PubMed] [Google Scholar]
- Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, Bremer RC, Rosenberg PS, & Savage SA (2016). Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute li-Fraumeni syndrome cohort. Cancer, 122(23), 3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabelli M, Cheng SC, & Parmigiani G (2016). Penetrance of ATM gene mutations in breast cancer: A meta-analysis of different measures of risk. Genetic Epidemiology, 40(5), 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Wubbenhorst B, D’Andrea K, Garman B, Long JM, Powers J, et al. (2014). Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genetics in Medicine, 17(8), 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocci E, Milne RL, Méndez-Villamil EY, Hopper JL, John EM, Andrulis IL, et al. (2013). Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiology and Prevention Biomarkers, 22(5), 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plichta JK, Griffin M, Thakuria J, & Hughes KS (2016). What’s new in genetic testing for cancer susceptibility? Oncology, 30(9), 787–799. [PubMed] [Google Scholar]
- Portnoy DB, Roter D, & Erby LH (2010). The role of numeracy on client knowledge in BRCA genetic counseling. Patient Education and Counseling, 81(1), 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017). R: A language for statistical computing. R Foundation for Statistical Computing, https://www.R-project.org/. [Google Scholar]
- Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward J, Philpott S, Anderson C, Edlund CK, Conti D, Harrington P, Barrowdale D, Bowtell DD, Alsop K, Mitchell G, AOCS Study Group, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Sieh W, McGuire V, Lester J, Bogdanova N, Dürst M, Hillemanns P, Ovarian Cancer Association Consortium, Odunsi K, Whittemore AS, Karlan BY, Dörk T, Goode EL, Menon U, Jacobs IJ, Antoniou AC, Pharoah PD, & Gayther SA (2015). Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. JNCI: Journal of the National Cancer Institute, 107(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Friebel T, Lynch HT, Neuhausen SL, van’t Veer L, Garber JE, et al. (2004). Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: The PROSE Study Group. Journal of Clinical Oncology, 22(6), 1055–1062. [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF, Antoniou AC, Nathanson KL, CIMBA Consortium, Laitman Y, Kushnir A, Paluch-Shimon S, Berger R, Zidan J, Friedman E, Ehrencrona H, Stenmark-Askmalm M, Einbeigi Z, Loman N, Harbst K, Rantala J, Melin B, Huo D, Olopade OI, Seldon J, Ganz PA, Nussbaum RL, Chan SB, Odunsi K, Gayther SA, Domchek SM, Arun BK, Lu KH, Mitchell G, Karlan BY, Walsh C, Lester J, Godwin AK, Pathak H, Ross E, Daly MB, Whittemore AS, John EM, Miron A, Terry MB, Chung WK, Goldgar DE, Buys SS, Janavicius R, Tihomirova L, Tung N, Dorfling CM, van Rensburg E, Steele L, Neuhausen SL, Ding YC, Ejlertsen B, Gerdes AM, Tv Hansen, Ramón y Cajal T, Osorio A, Benitez J, Godino J, Tejada MI, Duran M, Weitzel JN, Bobolis KA, Sand SR, Fontaine A, Savarese A, Pasini B, Peissel B, Bonanni B, Zaffaroni D, Vignolo-Lutati F, Scuvera G, Giannini G, Bernard L, Genuardi M, Radice P, Dolcetti R, Manoukian S, Pensotti V, Gismondi V, Yannoukakos D, Fostira F, Garber J, Torres D, Rashid MU, Hamann U, Peock S, Frost D, Platte R, Evans DG, Eeles R, Davidson R, Eccles D, Cole T, Cook J, Brewer C, Hodgson S, Morrison PJ, Walker L, Porteous ME, Kennedy MJ, Izatt L, Adlard J, Donaldson A, Ellis S, Sharma P, Schmutzler RK, Wappenschmidt B, Becker A, Rhiem K, Hahnen E, Engel C, Meindl A, Engert S, Ditsch N, Arnold N, Plendl HJ, Mundhenke C, Niederacher D, Fleisch M, Sutter C, Bartram CR, Dikow N, Wang-Gohrke S, Gadzicki D, Steinemann D, Kast K, Beer M, Varon-Mateeva R, Gehrig A, Weber BH, Stoppa-Lyonnet D, Sinilnikova OM, Mazoyer S, Houdayer C, Belotti M, Gauthier-Villars M, Damiola F, Boutry-Kryza N, Lasset C, Sobol H, Peyrat JP, Muller D, Fricker JP, Collonge-Rame MA, Mortemousque I, Nogues C, Rouleau E, Isaacs C, de Paepe A, Poppe B, Claes K, de Leeneer K, Piedmonte M, Rodriguez G, Wakely K, Boggess J, Blank SV, Basil J, Azodi M, Phillips KA, Caldes T, de la Hoya M, Romero A, Nevanlinna H, Aittomäki K, van der Hout A, Hogervorst FB, Verhoef S, Collée JM, Seynaeve C, Oosterwijk JC, Gille JJ, Wijnen JT, Gómez Garcia EB, Kets CM, Ausems MG, Aalfs CM, Devilee P, Mensenkamp AR, Kwong A, Olah E, Papp J, Diez O, Lazaro C, Darder E, Blanco I, Salinas M, Jakubowska A, Lubinski J, Gronwald J, Jaworska-Bieniek K, Durda K, Sukiennicki G, Huzarski T, Byrski T, Cybulski C, Toloczko-Grabarek A, Złowocka-Perłowska E, Menkiszak J, Arason A, Barkardottir RB, Simard J, Laframboise R, Montagna M, Agata S, Alducci E, Peixoto A, Teixeira MR, Spurdle AB, Lee MH, Park SK, Kim SW, Friebel TM, Couch FJ, Lindor NM, Pankratz VS, Guidugli L, Wang X, Tischkowitz M, Foretova L, Vijai J, Offit K, Robson M, Rau-Murthy R, Kauff N, Fink-Retter A, Singer CF, Rappaport C, Gschwantler-Kaulich D, Pfeiler G, Tea MK, Berger A, Greene MH, Mai PL, Imyanitov EN, Toland AE, Senter L, Bojesen A, Pedersen IS, Skytte AB, Sunde L, Thomassen M, Moeller ST, Kruse TA, Jensen UB, Caligo MA, Aretini P, Teo SH, Selkirk CG, Hulick PJ, & Andrulis I (2015). Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA, 313(13), 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendle KA, Halley MC, May SG, & Frosch DL (2015). Redefining risk and benefit: Understanding the decision to undergo contralateral prophylactic mastectomy. Qualitative Health Research, 25(9), 1251–1259. [DOI] [PubMed] [Google Scholar]
- Resta N, Pierannunzio D, Lenato GM, Stella A, Capocaccia R, Bagnulo R, Lastella P, Susca FC, Bozzao C, Loconte DC, Sabbà C, Urso E, Sala P, Fornasarig M, Grammatico P, Piepoli A, Host C, Turchetti D, Viel A, Memo L, Giunti L, Stigliano V, Varesco L, Bertario L, Genuardi M, Lucci Cordisco E, Tibiletti MG, di Gregorio C, Andriulli A, Ponz de Leon M, & AIFEG. (2013). Cancer risk associated with STK11/LKB1 germline mutations in Peutz–Jeghers syndrome patients: Results of an Italian multicenter study. Digestive and Liver Disease, 45(7), 606–611. [DOI] [PubMed] [Google Scholar]
- Riley BD, Culver JO, Skrzynia C, Senter LA, Peters JA, Costalas JW, Callif-Daley F, Grumet SC, Hunt KS, Nagy RS, McKinnon WC, Petrucelli NM, Bennett RL, & Trepanier AM (2012). Essential elements of genetic cancer risk assessment, counseling, and testing: Updated recommendations of the National Society of Genetic Counselors. Journal of Genetic Counseling, 21(2), 151–161. [DOI] [PubMed] [Google Scholar]
- Roed Nielsen H, Petersen J, Therkildsen C, Skytte AB, & Nilbert M (2016). Increased risk of male cancer and identification of a potential prostate cancer cluster region in BRCA2. Acta Oncologica, 55(1), 38–44. [DOI] [PubMed] [Google Scholar]
- Schmidt MK, Hogervorst F, Van Hien R, Cornelissen S, Broeks A, Adank MA, et al. (2016). Age-and tumor subtype–specific breast cancer risk estimates for CHEK2* 1100delC carriers. Journal of Clinical Oncology, 34(23), 2750–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim I, Gorman P, Greenes RA, Haynes RB, Kaplan B, Lehmann H, & Tang PC (2001). Clinical decision support systems for the practice of evidence-based medicine. Journal of the American Medical Informatics Association, 8(6), 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, Edlund CK, Conti D, Harrington P, Fraser L, Philpott S, Anderson C, Rosenthal A, Gentry-Maharaj A, Bowtell DD, Alsop K, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Høgdall E, Høgdall CK, Jensen A, Kjaer SK, Lubiński J, Huzarski T, Jakubowska A, Gronwald J, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Odunsi K, Goode EL, Menon U, Jacobs IJ, Gayther SA, & Pharoah PDP (2015). Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. Journal of Clinical Oncology, 33(26), 2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. (2017). Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the Clinical Genome Resource. The American Journal of Human Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, & Eng C (2012). Lifetime cancer risks in individuals with germline PTEN mutations. Clinical Cancer Research, 18(2), 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Broeke SW, Brohet RM, Tops CM, van der Klift HM, Velthuizen ME, Bernstein I, et al. (2014). Lynch syndrome caused by germline PMS2 mutations: Delineating the cancer risk. Journal of Clinical Oncology, 33(4), 319–325. [DOI] [PubMed] [Google Scholar]
- Theodoratou E, Campbell H, Tenesa A, Houlston R, Webb E, Lubbe S, Broderick P, Gallinger S, Croitoru EM, Jenkins MA, Win AK, Cleary SP, Koessler T, Pharoah PD, Küry S, Bézieau S, Buecher B, Ellis NA, Peterlongo P, Offit K, Aaltonen LA, Enholm S, Lindblom A, Zhou XL, Tomlinson IP, Moreno V, Blanco I, Capellà G, Barnetson R, Porteous ME, Dunlop MG, & Farrington SM (2010). A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. British Journal of Cancer, 103(12), 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton D, & Breast Cancer Linkage Consortium. (2001). Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. The American Journal of Human Genetics, 68(2), 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trialists’ Group ATAC (2005). Results of the ATAC (arimidex, tamoxifen, alone or in combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. The Lancet, 365(9453), 60–62. [DOI] [PubMed] [Google Scholar]
- Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, Timms K, Garber JE, Herold C, Ellisen L, Krejdovsky J, DeLeonardis K, Sedgwick K, Soltis K, Roa B, Wenstrup RJ, & Hartman AR (2015). Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer, 121(1), 25–33. [DOI] [PubMed] [Google Scholar]
- Tung N, Domchek SM, Stadler Z, Nathanson KL, Couch F, Garber JE, Offit K, & Robson ME (2016a). Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nature Reviews Clinical Oncology, 13(9), 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, Hartman AR, Winer EP, & Garber JE (2016b). Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. Journal of Clinical Oncology, 34(13), 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lier MGF, Wagner A, Mathus-Vliegen EMH, Kuipers EJ, Steyerberg EW, & Van Leerdam ME (2010). High cancer risk in Peutz–Jeghers syndrome: A systematic review and surveillance recommendations. The American Journal of Gastroenterology, 105(6), 1258–1264. [DOI] [PubMed] [Google Scholar]
- Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, Roeb W, Agnew KJ, Stray SM, Wickramanayake A, Norquist B, Pennington KP, Garcia RL, King MC, & Swisher EM (2011). Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proceedings of the National Academy of Sciences, 108(44), 18032–18037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dai B, & Ye D (2015). CHEK2 mutation and risk of prostate cancer: A systematic review and meta-analysis. International Journal of Clinical and Experimental Medicine, 8(9), 15708–15715. [PMC free article] [PubMed] [Google Scholar]
- Win AK, Reece JC, Dowty JG, Buchanan DD, Clendenning M, Rosty C, Southey MC, Young JP, Cleary SP, Kim H, Cotterchio M, Macrae FA, Tucker KM, Baron JA, Burnett T, le Marchand L, Casey G, Haile RW, Newcomb PA, Thibodeau SN, Hopper JL, Gallinger S, Winship IM, Lindor NM, & Jenkins MA (2016). Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. International Journal of Cancer, 139(7), 1557–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, Yang LS, Huang F, Hao JH, Su PY, & Sun YH (2012). Current evidence on the relationship between two polymorphisms in the NBS1 gene and breast cancer risk: A meta-analysis. Asian Pacific Journal of Cancer Prevention, 13(11), 5375–5379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.