Abstract
Since mammalian tissue is relatively transparent to near-infrared (NIR) light, NIR fluorescent proteins (FPs) engineered from bacterial phytochromes have become widely used probes for non-invasive in vivo imaging. Recently, these genetically encoded NIR probes have been substantially improved, enabling imaging experiments that were not possible before. Here, we discuss the use of monomeric NIR FPs and NIR biosensors for multiplexed imaging with common visible GFP-based probes and blue light-activatable optogenetic tools. These NIR probes are suitable for visualization of functional activities from molecular to organismal levels. In combination with advanced imaging techniques, such as two-photon microscopy with adaptive optics, photoacoustic tomography and its recent modification, reversibly switchable photoacoustic computed tomography, NIR probes allow subcellular resolution at millimeter depths.
Keywords: iRFP, biosensor, deep-tissue imaging, all-optical electrophysiology, bacterial phytochrome
Monomeric NIR FPs and biosensors open new possibilities for imaging and optogenetics.
Near-infrared fluorescent proteins (FPs) derived from bacterial phytochrome photoreceptors are useful probes for imaging across spatial scales from subcellular to the whole-body. In microscopy, NIR FPs allow spectral multiplexing with FPs of green fluorescent protein (GFP) family and optogenetic (see Glossary) tools activated by blue light. In deep-tissue imaging, deep penetration of NIR light in tissue, low autofluorescence and reduced scattering in NIR make NIR FPs superior than GFP-like FPs (reviewed in [1]). Therefore, NIR FPs have been widely used in various fields in biology and biomedicine, including cancer research [2–7], neuroscience [8–10], stem cell studies [11–13], parasitology [14], virology [15], where the probes’ NIR spectra allow their non-invasive visualization in vivo or labeling of organelles and cells for multicolor imaging.
Most NIR FPs are engineered chromophores that bind the PAS and GAF domains of bacterial phytochromes. As chromophores, bacterial phytochromes utilize biliverdin (BV), which is available in mammalian cells as a product of heme breakdown. Currently, there are about twenty different NIR FPs developed from bacterial phytochromes that can be grouped in series named by their developers: iRFPs [16, 17] and miRFPs [18, 19], IFPs [20, 21] and mIFP [22], and Wi-Phy [23], m/iRFP and m/IFP series, which were tested for their performance in mammalian cells, differ in their molecular brightness, effective brightness when expressed in mammalian cells (cellular brightness), spectral properties, and oligomeric state (Table 1, Key Table). A recent NIR FP developed from cyanobacterial phycobiliprotein photoreceptor smURFP [24] is considerably dimmer than iRFPs [25], although it is an interesting engineering result showing that a photoreceptor may be evolved to bind BV autocatalytically.
Table 1, Key Table.
BphP derived NIR FPs with demonstrated in vivo and live-cell applications and far-red GFP-like FPs.
| NIR FP | Ex, nm | Em, nm | Extinction coefficient, M−1cm−1 | Quantum yield, % | Molecular brightness a | Molecular brightness vs. iRFP713, % | Oligomeric state | Photostability in mammalian cells, t1/2, s | pKa | Brightness in HeLa cells vs. iRFP713, % b | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRFP670 | 642 | 670 | 87,400 | 14.0 | 12.2 | 198 | monomer | 490 (155) | 4.5 | 72 | [18] |
| miRFP703 | 674 | 703 | 90,900 | 8.6 | 7.8 | 127 | 650 (394) | 4.5 | 37 | ||
| miRFP709 | 683 | 709 | 78,400 | 5.4 | 4.2 | 69 | 500 (192) | 4.5 | 30 | ||
| mIFP bc | 683 (683) | 705 (704) | 65,900 (82,000) | 6.9 (8.4) | 4.6 | 74 | 90 (54) | 4.5 | 15 | [22] | |
| IFP2.0 bc | 688 (690) | 709 (711) | 72,900 (98,000) | 6.8 (8.1) | 5.0 | 80 | dimer d | 150 (108) | 4.5 | 8 | [21, 22] |
| iRFP670 | 643 | 670 | 114,000 | 12.2 | 13.9 | 225 | dimer | 290 | 4.5 | 119 | [17, 21] |
| iRFP682 | 663 | 682 | 90,000 | 11.1 | 10.0 | 162 | 490 | 4.5 | 105 | ||
| iRFP702 | 673 | 702 | 93,000 | 8.2 | 7.6 | 124 | 630 | 4.5 | 61 | ||
| iRFP713 (aka iRFP) | 690 | 713 | 98,000 | 6.3 | 6.2 | 100 | 960 | 4.5 | 100 | [16] | |
| iRFP720 | 702 | 720 | 96,000 | 6.0 | 5.8 | 93 | 490 | 4.5 | 112 | [16, 17] | |
| miRFP670–2 | 643 | 670 | 103,000 | 13.6 | 14.0 | 227 | monomer | 310 | 4.5 | 72 | [29] |
| miRFP682 | 663 | 682 | 91,000 | 11.2 | 10.2 | 165 | 500 | 4.5 | 117 | ||
| miRFP702 | 673 | 702 | 88,000 | 8.1 | 7.1 | 115 | 640 | 4.5 | 37 | ||
| miRFP713 | 690 | 713 | 99,000 | 7.0 | 6.9 | 112 | 980 | 4.5 | 109 | ||
| miRFP720 | 702 | 720 | 98,000 | 6.1 | 6.0 | 97 | 510 | 4.5 | 116 | [19] | |
| mCherrye | 587 | 610 | 72,000 | 22.0 | 15.8 | 255 | monomer | n.a. | 3.8 | n.a. | [19, 28] |
| FusionRede | 580 | 608 | 83,000 | 19.0 | 15.8 | 255 | n.a. | 4.6 | n.a. | [28, 37] | |
| mNeptunee | 600 | 650 | 57,500 | 20.0 | 11.5 | 185 | dimer f | n.a. | 5.4 | n.a. | [37, 72, 73] |
| mCardinale | 603 | 651 | 79,000 | 18.0 | 14.2 | 229 | n.a. | 5.3 | n.a. | [72–74] |
Determined as a product of extinction coefficient at excitation maximum (in mM−1cm−1) and fluorescence quantum yield.
Determined as an effective NIR fluorescence in transiently transfected live HeLa cells with no supply of exogenous BV and after normalization to fluorescence of co-transfected EGFP and overlap of FP spectra with excitation laser and emission filters.
Characteristics of NIR FPs shown in original publications are in parentheses.
Originally reported as a monomer, IFP2.0 was later found to be a dimer [22]
Farred GFP-like FP.
Originally reported as monomers, mNeptune and mCardinal were later found to be dimers [73].
n.a., not available.
Relatively low quantum yield of NIR FPs is compensated by their high extinction coefficient. The resulting molecular brightness of NIR FPs is comparable to modem far-red FPs of green fluorescent protein (GFP) family (Table 1). NIR FPs are used in microscopy in the same constructs and imaging conditions as GFP-like probes. However, it is important to use light sources that produce adequate light intensities in NIR, like xenon lamps. In addition, cellular and tissue autofluorescence is lowest in the NIR spectral region [4, 26], In deep-tissue imaging, iRFPs were shown to be superior than brightest available far-red GFP-like FPs in direct comparison [16, 17], due to low autofluorescence in NIR combined with better NIR light penetration and less scattering. Interestingly, relatively dim and the most red-shifted iRFP720 was found to be the most sensitive probe among FPs for deep-tissue imaging [15, 27].
Apparent brightness of NIR FPs in cells, so called cellular brightness, does not always correlate with its molecular brightness [1, 25], It also depends on protein stability and BV binding efficiency. Early NIR FPs, such as IFP1.4 [20], suffered from poor BV incorporation in mammalian cells leading to their poor brightness (8% for IFP1.4 vs iRFP713 [17]). Screening protein variants in mammalian cells during protein engineering allowed to obtain iRFPs that do not require exogenous BV supply and can be used similarly to GFP-like probes [16, 17].
Bright NIR FPs, such as the iRFP family, are dimers and may not be suitable for protein tagging and engineering of biosensors as they can interfere with proper localization and functions of fused functional molecules by inducing their dimerization. Recent development of bright monomeric NIR FPs [18, 19, 22] has changed the situation the same way that the development of monomeric mFruits [28] from dimeric DsRed GFP-like FPs did a decade ago. Moreover, with advances in technical approaches, NIR probes can allow deeper in vivo imaging at higher resolution than available before. Here we focus on new possibilities opened by monomeric NIR FPs and biosensors. We overview the monomeric NIR FPs and their properties, describe uses of NIR probes for multiplexed experiments, discuss the newly designed NIR Förster resonance energy transfer (FRET) pair and NIR FRET biosensors, describe NIR reporters based on changes in protein amount and their use across spatial scales, and provide examples where synergistic advancements in probe engineering and development of imaging techniques resulted in resolution as high as subcellular at mm depths in tissue.
A palette of monomeric NIR FPs
Recently developed monomeric miRFPs have properties similar to dimeric iRFPs (Table 1); however, the monomeric state makes them suitable for a variety of protein fusions. An engineering challenge was involved in developing bright monomeric FPs: simple disruption of the dimeric interface known from the crystal structures results in decreased brightness in mammalian cells, possibly caused by reduced protein stability. The first bright monomeric NIR FPs—mIFP [22], and miRFP670, miRFP703, and miRFP709 [18]—were engineered from bacterial phytochromes that do not dimerize via their chromophore-binding domain. Additional mutagenesis on the possible dimerizing interface in miRFPs resulted in FPs that behaved as monomers even at high concentration of 30 mg/ml. Building on this result and structure studies, dimeric iRFPs were monomerized without a loss of brightness (Fig. 1A) [19, 29], These FPs have properties (Table 1) similar to their dimeric counterparts and make a whole iRFP palette available as miRFPs. Importantly, miRFPs inherited high effective cellular brightness of dimeric iRFPs resulting from efficient BV incorporation. Monomeric state enabled a use of miRFPs as versatile fusion tags. It was shown that miRFPs indeed localize well in protein fusions (Fig. 1B).
Figure 1. NIR FPs and their applications in multiplexed imaging and optogenetics.
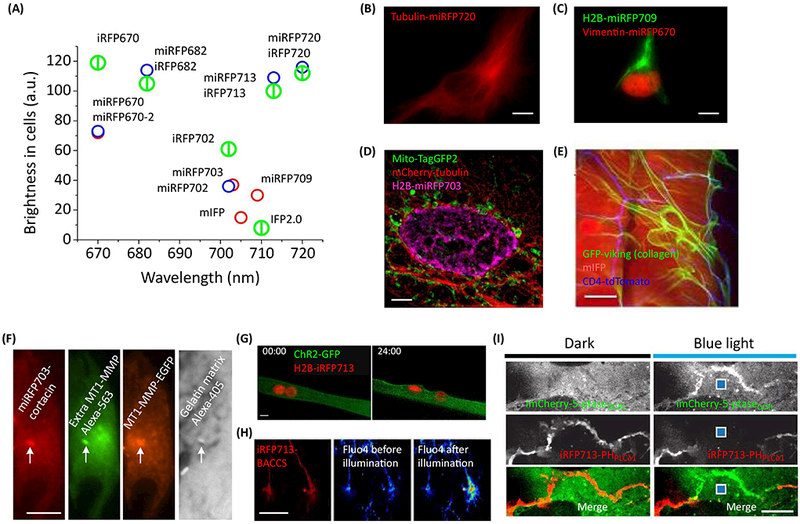
(A) Effective cellular brightness and emission wavelength maxima of NIR FPs. Early miRFPs and mIFP are shown as red circles, new miRFPs as blue circles, and dimeric iRFPs as larger green splitted circles. (B) Tubulin-miRFP720 localizes well in live HeLa cells. (C) miRFP670 and miRFP709 allow two-color labeling as shown by simultaneous visualization of H2B and vimentin in HeLa cells. (D, E) miRFPs and mIFP allow cross-talk free three-color imaging together with green and red GFP-like FPs as shown by structured illumination microscopy (SIM) in HeLa (D), and by visualization of abdomen muscle (mIFP), Class IV DA neurons (CD4-tdTomato), and extracellular collagen matrix (GFP) in Drosophila (E). C, D adapted under the Creative Commons Attribution license from [18]; E adapted with permission from [22], (F) miRFPs was used in four- color imaging to localize invadopod (marked by white arrow, miRFP703), colocolized with intracellular (EGFP) and extracellular (Alexa-565) MT1-matrix metalloproteinase (MMP), and matrix degradation (Alexa-405). Courtesy of Louis Hodgson (G-I) iRFP713 was used with blue-light induced optogenetic tools, including chanelrhodopsin ChR2 in muscle (G, adapted with permission from [33]), channel switch BACCS in neurons (H, adapted under the Creative Commons Attribution license from [34]) and system CRY2/CIBN for recruitment of inositol-5-phosphatase (5-ptase) to the cell membrane in COS-7 cells (I, adapted with permission from [35]). Scale bars, 5 μm (D, I), 10 μm (B, C, G), 20 μm (E, F), and 50 μm (H).
Multiplexed imaging and optogenetics with NIR genetically encoded probes
NIR probes complement GFP-like probes for multicolor imaging. Moreover, spectrally distinct m/iRFPs can be used in multicolor imaging (Fig. 1C). miRFPs and mIFP allow cross-talk free multicolor labeling with green and red GFP-like FPs (Fig. ID, E). miRFP703 was used in multicolor imaging in a study of a mechanism of invasion and metastasis in cancer cells associated with activity of Rac3 GTPase localized at invadopodia [30], Invadopodia, which are plasma membrane protrusions, mediate the degradation of extracellular matrix promoting cancer cell invasion. Four-color imaging of miRFP703-labeled invadopod core protein cortactin was used to detect mature invadopodia that colocalized with intracellular and extracellular-exposed MT1-matrix metalloproteinase (MMP) responsible for matrix degradation (Fig. 1F). Cortactin-miRFP703 was also used to visualize invadopodia simultaneously with the activity of Rac3 biosensor based on mCerulean-mVenus FRET pair [30], Dimeric iRFPs can be used for multicolor imaging to highlight a whole cell or organelle, and in protein fusions where their dimeric state does not interfere with functionality (see examples in [31] for multicolor tracing of motor recruitment to cargo and in [32] for use of iRFP713 in AAVs in mice expressing EGFP and tdTomato).
The NIR spectra of iRFPs make them ideal probes for multiplexing with blue-green optogenetic tools. For example, an iRFP713 was used to track nucleus movement to the periphery in muscle cells after optogenetically (with channelrhodopsin ChR2) induced contraction (Fig. 1G) [33], Also, iRFP713 was used to label an engineered blue light-activated calcium channel switch named BACCS [34], BACCS activation results in calcium-induced gene expression. Changes in the calcium concentration were detected with green Fluo4/AM indicator simultaneously with visualization of NIR iRFP713-BACCS (Fig. 1H). Another example is a use of iR.FP713-PHPLCδ1 fusion as a biosensor for blue-light induced dephosphorylation of lipid phosphoinositide PI(4,5)P2 via engineered optogenetic system (Fig. II) [35], Optogenetic control of lipid modifications coupled with visualization of these modifications is an example of a powerful all-optical approach to study cell signaling in their native environment. iRFP713 was furthermore used to label optogenetic channels allowing their visualization without affecting opsin functions [9], Although these and other experiments were possible with dimeric iRFPs, the availability of miRFPs should make multiplexed imaging and optogenetics widely applicable, because monomeric miRFPs are versatile protein labels and optimal building blocks for engineering of various biosensors for cell signaling pathways.
Fully NIR FRET pair and FRET-based biosensors
Engineering multispectral monomeric miRFPs allows designing FRET pairs with versatile uses. Earlier FRET pair of dimeric iRFP713 combined with mKate2 was suboptimal and applied only to caspase-3 sensor [36], Possibly, a combination of miRFPs with advanced red fluorescent proteins (RFPs) [37, 38] can produce an efficient FRET pair with high dynamic range of response. Recently a fully NIR FRET pair was reported [19], Advantages of NIR FRET over RFP-NIR FP FRET are similar to advantages of NIR FPs over RFPs: better spectral separation in multispectral experiments, lower autofluorescence background, less scattering and better light penetration in tissue for in vivo experiments. miRFP670-miRFP720 FRET pair is characterized by good spectral overlap between the donor emission and the acceptor excitation, minimal direct cross-excitation of the acceptor among miRFP combinations, and a high Förster radius (Ro) of 8.3 nm [19], For comparison, Ro is 4.9 nm for ECFP-EYFP and 5.4 nm for mCerulean-mVenus [39], The performance miRFP670-miRFP720 FRET pair was tested in a fusion containing caspase-3 cleavage site. The response of 34% changes in the donor/FRET ratio for miRFP670-miRFP720 is comparable to responses of cyan-yellow CFP-YFP FRET pairs [40, 41], Therefore, this NIR FRET pair can be used to re-design FRET biosensors into the NIR spectral range.
GFP-like FRET biosensors for key molecules in cellular signaling including Ca2+, cyclic AMP, phospholipids, small GTPases, protein kinases are widely used in studies of signaling cascades. NIR FRET biosensors for these molecules are in high demand and will be engineered in the near future. By exchanging GFP-like FPs for miRFPs in kinase sensors [42], NIR biosensors for protein kinase A (PKA) AKAR and c-Jun N-terminal kinases (JNK) JNKAR were obtained (Fig. 2A-C). Further optimization by changing the length and composition of linkers can improve the response of the sensors, as was performed for the NIR biosensor for Racl GTPase, which regulates signaling pathways controlling cell motility [43] (Fig. 2D) [19], The resulting Racl biosensor is a robust probe with a high dynamic range of response (up to 2.7-fold changes in FRET/donor ratio). The NIR Racl sensor can be combined with CFP-YFP biosensors and blue-green optogenetic tools. Simultaneous imaging of Racl in NIR and other GTPases in visible range allows to directly visualize spatiotemporal coordination of GTPase activities at the cell leading edge that result in protrusion-retraction cycles and stalling of the edge. By imaging of NIR Racl and CFP-YFP RhoA biosensors, the antagonism between activities of these two GTPases was directly characterized in the same cell, for the first time (Fig. 2E). Spatiotemporal coordination of Racl activity by its negative regulator GDI was revealed by simultaneous imaging of NIR Racl and CFP-YFP Racl-GDI binding sensor. Further, this biosensor was used simultaneously with optogenetic activation of the Racl GTPase by the LOV-TRAP tool.
Figure 2. Single-chain NIR biosensors for different targets in signaling cascades based on miRFP670-miRFP720 FRET pair.
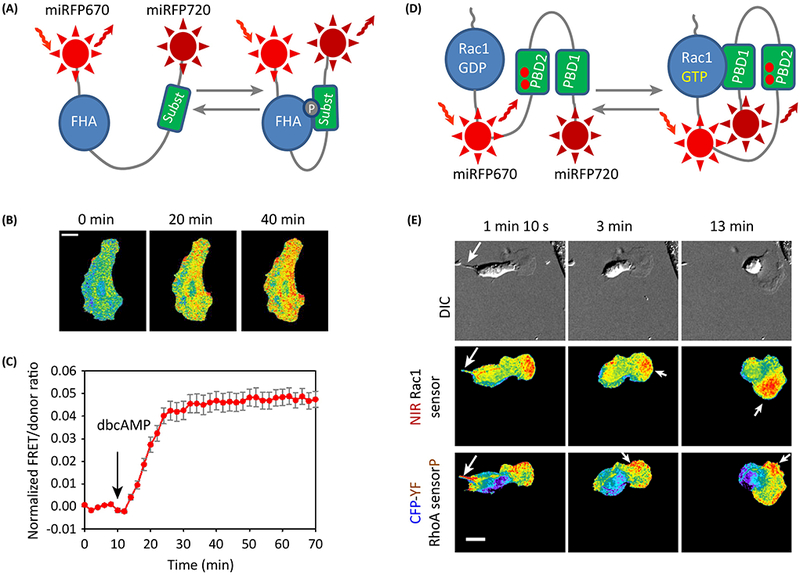
(A) Schematics of the NIR FRET biosensors for AKAR PKA and JNKAR JNK kinases. FHA is the phosphopeptide binding domain that recognizes phosphorylated peptide marked as “subst“. (B,C) NIR AKAR PKA kinase biosensor was validated in live HeLa cell stimulated with 1 mM dibutyryl cAMP (dbcAMP). Time-lapse images (B) and the corresponding plot (C) are shown. (D) Schematics of the FRET Racl biosensor. PBD1 is a p21-binding domain 1, PBD2 is a mutant p21-binding domain. Racl is a full-length Racl post-translationally isoprenylated for membrane localization. (E) Combination of NIR and cyan-yellow FRET biosensors for simultaneous imaging of Racl and RhoA activities in a MEF/3T3 cell. DIC, differential interference contrast. Racl is predominantly localized at the leading edge, whereas RhoA activity is mostly localized at the retracting tail, the side edges and at the back of the leading-edge protrusions (see arrows). (B,E) FRET/donor ratio is shown in pseudocolor. Scale bar, 20 μm. (C) Mean ±s.e.m. (n=3) of the FRET/donor ratio for the whole cell plotted vs time. B,C,E adapted with permission from [19]
NIR reporters for imaging across scales
A toolbox of NIR FRET and intesiometric biosensors based on conformational changes in the protein backbone upon activation is yet to be developed. NIR reporters based on changes in the amount of fluorescent tag because of its synthesis/degradation or reconstitution are already available and have been applied to imaging at different spatial scales.
The reporter for canonical activation of NF-κB pathway is a fusion of miRFP703 with IkBα, which binds NFκB transcription factor and keeps it in the cytoplasm [18], This pathway is associated with immune response, cellular proliferation and apoptosis [44], Upon its activation, IkBα is degraded together with miRFP703 and decrease of fluorescence can be detected (Fig. 3 A). This reporter was visualized in cells upon addition of TNFα (Fig. 3B), and in animals upon injection of lypopolysachride (LPS) triggering inflammation (Fig. 3C).
Figure 3. Multiscale imaging of NIR reporters.
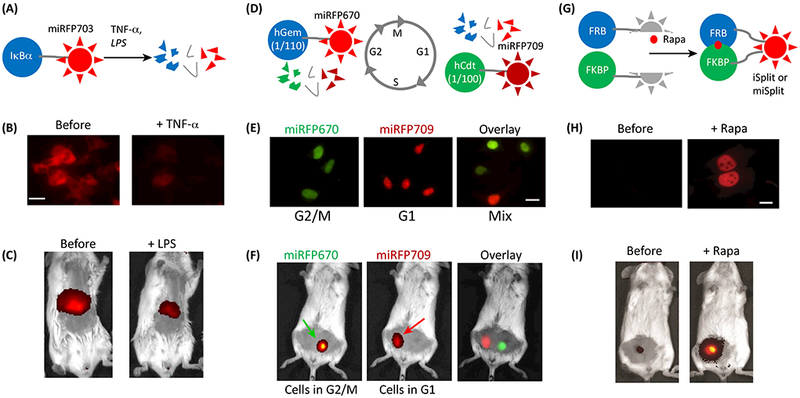
(A) Schematics of the IκBa-miRFP703 reporter for canonical activation of NF-κB pathway. Its activation by TNFα or LPS results in hcBa degradation together with miRFP703. (B,C) Visualization of IκBa-miRFP703 reporter in HEK293 cells and in a mouse liver. B,C adapted under the Creative Commons Attribution license from [18], (D) Schematics of the NIR cell cycle reporter. miRFP670-hGem(l/110) and miRFP709-hCdtl(l/100) are reciprocally produced and degraded in a cell cycle dependent manner. (E,F) Visualization of NIR cell cycle reporter in synchronized (in G2/M and G1 phases) and non-synchronized (mix) HeLa cells and in a mouse with implanted cells. E,F adapted under the Creative Commons Attribution license from [18], (G) Schematics of the NIR bimolecular fluorescence complementation reporters iSplit and miSplits. Two split fragments (the PAS and the GAF domains of iRFP or miRFP) are not fluorescent as separate polypeptides until two fusion proteins interact. Here FRB and FKBP interact in the presence of rapamycin (rapa). (H,I) Visualization of iSplit reporter in HeLa cells and in a mouse xenograft tumor. H,I adapted with permission from [47].
The NIR cell cycle reporter [18] is based on Fucci technology [45], which consists of two spectrally distinguishable miRFP670 and miRFP709 fused to the fragments of Geminin and Cdt1. The levels of these proteins participating in the licensing of replication origins change reciprocally (Fig. 3D). Using this reporter, dividing cells were distinguished from non-dividing in culture (Fig. 3E) and implanted in mice (Fig. 3F) by detecting fluorescence in two channels corresponding to miRFP670 and miRFP709. Similar cell cycle reporter was developed using a pair of IFP2.0 and smURFP [24].
NIR reporters for protein-protein interactions are based on bimolecular fluorescence complementation (BiFC) of the two split fragments: the PAS and the GAF domains of NIR FPs (Fig. 3G) [46–49]. An iSplit reporter based on iRFP713 was successfully applied in mammalian cells (Fig. 3H) and in mice (Fig. 3I) [47]. New monomeric versions miSplit709 and miSplit670 have several fold lower background and higher contrast than iSplit [18]. BiFC contrast decreases if split fragments tend to self-associate. BiFC contrast in cultured cells for miSplits was found to be 18-fold for miSplit709 and 41-fold for miSplit670 [18]. For comparison, the highest reported contrast for BiFC derived from GFP-like Venus in similar conditions was reported to be 17-fold [50]. miSplit670 and miSplit709 share the same PAS domain, but differ in their GAF domains, specifically, amino acids near the chromophore in the GAF domains that define spectral properties. Thus, the combination of miSplit670 and miSplit709 can be adapted for detection of interaction of the protein fused to the PAS domain with two alternative binding partners fused to the mGAF670 or mGAF709 domains by the wavelength of the reconstituted fluorescence [18]. miSplits were also applied to RNA labeling. Fused to two high-affinity bacteriophage RNA-binding proteins MCP and PCP, miSplits reconstituted fluorescence when bound to mRNA molecules tagged with 12 repeats of corresponding RNA-binding sites (MBS and PBS). Since self-assembly for miSplits and the autofluorescence background are low, this techniques provides a sensitive method for RNA detection.
Circular permutation of IFP proteins was used to make a fluorogenic reporter for protease activity, including iCasper applied to image apoptosis during morphogenesis and tumorigenesis in Drosophila [51]. This reporter is based on cleavage-dependent chromophore incorporation.
Scaling down in spatial resolution, miRFPs and mIFP probes were applied to subdiffraction structured illumination microscopy (SIM) (Fig. 1D) [18, 22]. Further, NIR FPs were used in recent two-wavelength total internal reflection fluorescence (TIRF) microscopy STAR (simultaneous two-wavelength axial ratiometry) with 20 nm axial resolution [52]. Using this method, endocytosis of EGFR receptor labeled with both EGFP and iRFP713 was visualized in real time.
Imaging of NIR probes in vivo with enhanced resolution
Light scattering, aberrations, tissue absorbance, and autofluorescence are major obstacles for in vivo imaging. Deep penetration of NIR light in tissue and low autofluorescence in NIR make NIR FPs suitable for sensitive imaging in vivo (Table 2). However the resolution is poor due to light scattering, which is reduced in NIR but still present, and optical aberrations. In this section, we discuss recent imaging techniques that use NIR probes to obtain higher resolution at larger depths.
Table 2.
Methods for imaging of NIR probes across spatial scales.
| Method | Probe | Spatial resolution | Temporal resolution | Sensitivity | Spectral multiplexing | Refer ence |
|---|---|---|---|---|---|---|
| Animal whole-body, deep tissues and organs imaging | ||||||
| Planar fluorescence imaging | iRFPs, miRFP. NIR biosensors | >1 mm up to 2 cm deep | >1 ms time resolution* | >105 cells up to 2 cm deep | Two color imaging with m/iRFP670 and m/iRFP720; Combination with blue light optogenetic tools | [1, 15, 17] |
| Fluorescence tomography | >1 mm up to 2 cm deep | <1 s time resolution* | [2, 59] | |||
| Planar NIR bioluminescence imaging | iRFPs-RLuc8s | >1 mm up to 2 cm deep | >1 ms time resolution** | >104 cells up to 2 cm deep | [75] | |
| Fluorescence lifetime tomography | iRFPs, miRFP, NIR biosensors | ˜1 mm up to 1 cm deep | <1 s time resolution* | ˜104 cells up to 1 cm deep | [76] | |
| Photoacoustic tomography | <1 mm up to 1 cm deep | <1 s time resolution* | >104 cells up to 1 cm deep | [57–59,77] | ||
| Reversibly-switchable photoacoustic tomography | BphPl | >100 μm up to 1 cm deep | >1 s time resolution | >102 cells up to 1 cm deep | Combination with blue light optogenetic tools | [60,78] |
| In vivo cellular and subcellular imaging | ||||||
| Two-photon (2P) microscopy | iRFPs, miRFP, NIR biosensors | 10 μm up to 1 mm deep | >1 ms time resolution* | 1 cell up to 1 mm deep | Combination with blue light optogenetic tools | [10,55] |
| 2P microscopy with adaptive optics (AO) |
>1 μm up to 1 mm deep | Subcellular up to 1 mm deep | [56] | |||
| Ex vivo cell analysis | ||||||
| Flow cytometry | miRFPs, iRFPs | Individual cells | – | Individual cells | <4 multispectral m/iRFPs and several dyes/GFP-like FPs | [17,79] |
| Cells and subcellular structures | ||||||
| Fluorescence microscopy (wide-field and confocal) | iRFPs, miRFP, | >350 nm | >1 ms time resolution* | Molecules* | Cross-talk free imaging with green and red GFP-like FPs; |
[18,19,22] |
| Structured illumination | NIR biosensors | >175 nm | >1 ms time | Subcellular | multicolor imaging; Combination with blue | [18, 22 |
| microscopy (SIM) | resolution* | structures | light optogenetic tools | 22] | ||
Depends on the power of light source and sensitivity of the detector/camera.
Depends on camera sensitivity.
Two-photon (2P) microscopy is traditionally used for cellular and subcellular resolution in vivo imaging, especially in neuroscience [53]. Some properties of iRFPs in 2P microscopy have recently been characterized [10]. For example, a standard Ti-Sapphire laser excites iRFPs at 880 nm more effectively than the laser equipped with optical parametric oscillator (OPO) at 1280 nm (Fig. 4A). Thus, iRFPs are suitable for single wavelength two-photon imaging together with EGFP/EYFP at 880 nm (Fig. 4B). Further improvement of resolution at tissues located at depths up to 1.6 mm can be achieved by using a combination of adaptive optics (AO) with 2P microscopy [54, 55]. The reduced scattering of NIR fluorescence promoted the use of iRFPs as guide stars for correcting aberrations by applying direct wavefront sensing method [56]. Using this technique, iRFP713 optical guide-star and green calcium biosensor GCaMP6 were imaged in primary visual cortex of a mouse at depth of 550 gm with subcellular resolution (Fig. 4C).
Figure 4. High resolution in vivo imaging of NIR probes.
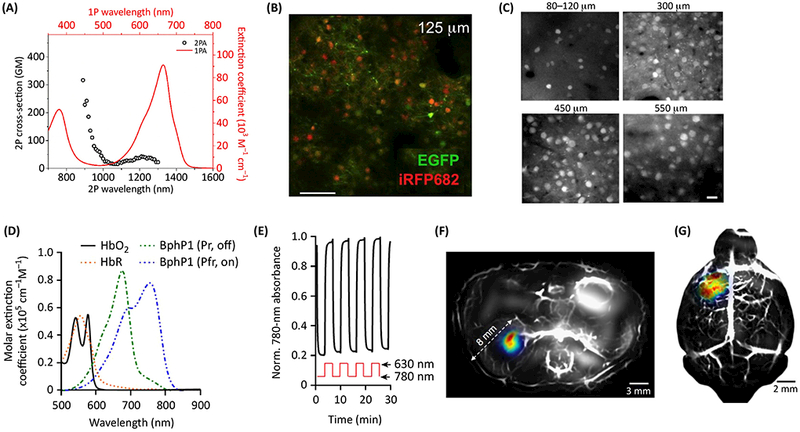
(A) iRFPs can be effectively excited by Ti-Sapphire laser at 880 nm as shown by two-photon (2P) absorption spectra of iRFP682 (open circles). One-photon (solid line) absorption spectra of iRFP682 is also shown. (B) Visualization of iRFP682 and EGFP in neurons of mouse cortex layer 2/3 using in vivo single-wavelength (880 nm) 2P microscopy. A,B adapted with permission from [10], (C) Visualization of iRFP713 in neurons of mouse cortex layers 1, 2/3, 4, 5 using 2P microscopy with adaptive optics wavefront correction. C adapted with permission from [9] and [56], (D) The photoacoustic tomography (PAT) of BphPl Pfr (marked as ON) and BphPl Pr (marked as OFF) allows to subtract background absorption by hemoglobin (Hb02) and deoxyhemoglobin (HbR). (E) BphPl can be repeatedly photoswitched by 780 and 630 nm light between its Pfr ON state and Pr OFF state. (F, G) Reversibly switchable photoacoustic computed tomography (RS-PACT) of deep-seated tumors in vivo. This technique can visualize kidney (F) and brain (G) tumors and vasculature as overlays of the ON state image (BphPl, pseudocolor) and the OFF state image (hemoglobin, gray) with exceptionally high resolution. F,G adapted with permission from [60], Scale bars, 100 μm (B), 20 μm (C).
Another approach to overcome the problem of light scattering in vivo is a photoacoustic tomography (PAT). iRFPs are genetically encoded probes of choice for PAT because their absorption is well differentiated from hemoglobin in blood. Their exceptional performance in PAT to visualize deep-seated tumors with submillimeter resolution at up to 8 mm depth has been extensively utilized [57–59]. A recent approach, called reversibly switchable photoacoustic computed tomography (RS-PACT), enabled even deeper imaging at higher resolution: 100 gm at depths up to 10 mm [60]. This technique relies on the reversibly photoswitchable bacterial phytochrome BphP1, which is a precursor of miRFPs (Fig. 4D,E) and uses differential imaging to cancel non-specific light absorption by tissue. Using this technique, small deep-seated tumors undetectable with traditional PAT in the kidney and in the brain were visualized (Fig. 4F,G).
Conclusions and future perspectives
The recent development of monomeric NIR FP, reporters and biosensors allow direct monitoring of active processes in multiplexed experiments at different spatial scales (Table 2). Combined with advanced imaging technologies, such as 2P microscopy with AO, and PAT, they should help to mitigate the current tradeoff between imaging depth, spatial resolution, and field of view (Fig. 5).
Figure 5. Optical technologies aided by NIR probes to address current challenges.
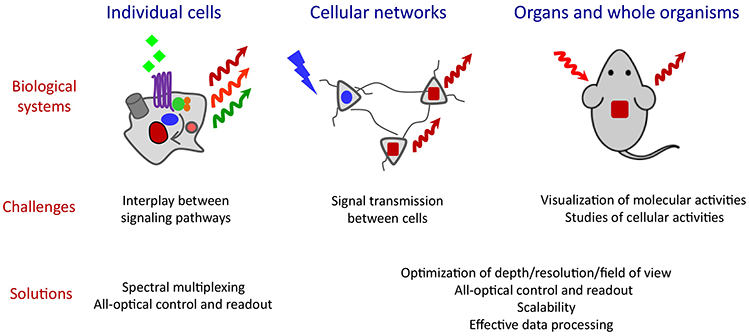
Challenging studies at different spatial scales from cells, cellular networks to organs and whole organisms (top, “Biological systems”) are presented in the “Current challenges” (middle). Corresponding optical technologies that rely on NIR FPs and NIR biosensors and could address the challenges are listed in the “Solutions” (bottom).
The brightest NIR FPs (Table 1), such as iRFPs and miRFPs, are sufficiently optimized and can be used as easily as GFP-like FPs. NIR FPs have low cytotoxicity and high photostability, but their quantum yield can be improved (see Outstanding Questions). Future protein engineering efforts may address this challenge by using structure insights [61, 62], novel precursor bacterial phytochromes for evolution, and advanced screening approaches, such as using fluorescence lifetime that is insensitive to expression level of fluorescent probe and thus allows for selection of bacteria expressing the intrinsically bright probes [63]. Another challenge is to the shift spectra of FPs further in NIR region to allow deeper light penetration in tissue, lower autofluorescence background and thus higher imaging sensitivity. Attempts to shift the spectra by stabilizing the Pfr state of phytochromes absorbing at ˜740 nm have not been successful so far.
The NIR Rac1 biosensor should inspire the generation of more NIR biosensors for a wide variety of cell signaling activities based on NIR FRET technology. The activities of interest include tyrosine and serine-threonine kinases; small GTPases; and second messengers including calcium, cAMP, and lipid-derived signaling molecules. Compatible with GFP-like probes and optogenetic tools, NIR biosensors should allow studies that are currently challenging, such as visualizing multiple events in signaling pathways and detecting interplay between pathways, studying signal transmission between cells, and performing visualization and causal studies of molecular and cellular activities in vivo (Figure 5).
Together with blue-green optogenetic tools, NIR biosensors should enable cross-talk free precise and patterned manipulation of cellular functions and simultaneous detection of molecular activities. Such all-optical control and readout can involve optogenetic tools to interrogate signaling cascades, starting from receptor activation and ending with gene transcription and genome editing (reviewed in [64, 65]). NIR biosensors for calcium and voltage changes are highly desired for combination with channelrhodopsins to allow all-optical electrophysiology [66]. Although this approach was applied to study neuronal circuits [67] and cardiac dynamics [68], cross-talk between tools, low brightness of current red-shifted indicators, and a need for a specialized experimental setup prevent its wide use in research community.
We also anticipate future engineering of intensiometric biosensors based on modulation of the fluorescence of a single NIR FP. Whereas FRET sensors allow quantitative readouts and efficient segregation of their signal from hemodynamic signal components, intensiometric sensors may provide higher dynamic ranges of response and require only a single bandwidth for imaging. These sensors can be developed by engineering circularly permuted monomeric NIR FPs fused to sensing domains or by using FPs with insertions containing sensing domains that reversibly modulate fluorescence upon ligand binding [69].
The applicability of NIR biosensors to imaging at different spatial scales, from subcellular to organismal, should facilitate workflows in drug screening from cells to whole animals and stimulate the development of novel models of diseases. Since several transgenic animals have been developed using iRFPs (Drosophila [22, 70], mice [71] and rats [9]), we expect transgenic mice expressing NIR biosensors to be generated in the near future. These mouse models should advance preclinical studies by facilitating drug screening and validation in vivo.
Highlights.
New monomeric NIR fluorescent probes (FPs) complement GFP-like FPs for cross-talk free imaging. Spectrally distinct versions of these FPs are available, ranging from 670 nm to 720 nm in emission maxima.
The first NIR FRET biosensor, which detects Racl GTPase, is compatible with simultaneous imaging with CFP-YFP based biosensors.
Because they have excitation and fluorescence close to or within the NIR window of tissue transparency (650 nm - 900 nm), NIR probes can be imaged across scales from subcellular to whole animals. Functional imaging is possible using cell signaling, cell cycle and protein-protein interaction reporters.
Spectral properties of NIR FPs combined with advanced imaging approaches, such as structured illumination, two-photon microscopy and photoacoustic tomography, enable subcellular resolution at mm depths.
Outstanding Questions.
Is it possible to engineer NIR FPs with increased quantum yield? What is the best strategy for directed molecular evolution to achieve this goal?
How to engineer NIR FPs shifted towards NIR part of the spectrum with emission maxima beyond 720 nm? Is it possible to do this by stabilizing Pfr-state of bacterial phytochromes?
When will NIR FRET biosensors be developed for functional molecules in cell signaling cascades, such as protein kinases, phosphatases, GTPases, second messengers? Will miRFP670-miRFP720 FRET pair be suitable for re-engineering available GFP-like FRET sensors into NIR? How will NIR FRET biosensors perform comparing to original GFP-like probes?
When will NIR biosensors for calcium and voltage changes be developed to allow straightforward all-optical electrophysiology for causal studies with precise spatiotemporal resolution? How will they compare to available GFP-like biosensors?
Can NIR single-color intensiometric biosensors be generated using NIR FP scaffold? What is the best strategy to achieve this: circular permutation or insertion of sensing domains into unperturbed NIR FPs?
Is it possible to improve resolution and/or sensitivity in in vivo imaging with NIR FPs?
When will transgenic mouse models expressing NIR FPs and biosensors be available for a wide community of researchers and companies focused on preclinical studies?
Acknowledgements
This work was supported by grants GM122567 and NS103573 from the US National Institutes of Health, ERC-2013-ADG-340233 from the EU 7th Framework Programme (FP7), 16-04-01515 from the Russian Foundation for Basic Research, and grant from the Molecular and Cell Biology Program of the Russian Academy of Sciences. We apologize to researchers whose work we could not quote here due to space limitation and focus of this review.
Glossary
- All-optical electrophysiology
an approach that combines light-induced perturbation (using channelrhodopsins as optogenetic tools) and optical readout (via genetically encoded sensors for membrane voltage or calcium) of neuronal activity.
- Adaptive optics (AO)
correction of aberrations in scattering tissue to enhance resolution of the optical imaging at depth. A direct wave front-sensing approach using fluorescence guide stars is a method to correct these aberrations and increase depth of high-resolution imaging.
- Bacterial phytochromes
multidomain photoreceptors that respond to near-infrared (NIR) light and incorporate the most red-shifted natural chromophore biliverdin (BV). They participate in microbe light-adaptive behavior and activate a biochemical response following light-induced chromophore isomerization and conformational changes in the protein molecule.
- Bimolecular fluorescence complementation (BiFC)
an approach of studying of protein-protein interaction based on the reconstruction of a fluorescent probe from split halves fused to the target proteins.
- Cellular brightness
effective fluorescence of a NIR FP in a living cell that depends on molecular brightness of the probe, protein expression level, the folding efficiency and stability of the probe, and affinity and specificity of the apoprotein to BV chromophore.
- Circular permutation
a rearrangement of the order of protein sequence by connecting the original N- and C- termini via peptide linkers and introducing the novel protein ends.
- c-Jun N-terminal kinases (JNK)
a family of mitogen-activated serine/threonine protein kinases that participate in signaling pathways in stress responses, including cytokines, ultraviolet irradiation, heat and osmotic shocks, proliferation, embryonic development, and apoptosis.
- Förster radius (Ro)
characterizes the theoretical efficiency of Förster resonance energy transfer (FRET) between light-emitting donor and light-absorbing acceptor molecules. Ro is the distance at which the energy transfer efficiency is 50%. It depends on the overlap between the donor emission and the acceptor absorbance spectra, fluorescence quantum yield of the donor, molar extinction coefficient of the acceptor, refractive index of the medium, donor emission wavelength and calculated for freely rotating chromophores.
- Förster resonance energy transfer (FRET)
a mechanism of non-radiative energy transfer from a donor fluorophore in its electronic excited state to an acceptor fluorophore through dipole-dipole coupling.
- Fucci
short for fluorescence ubiquitination-based cell cycle indicator, a technology that uses a genetically encoded two-color indicator system to analyze cell-cycle dynamics within a cell population.
- Optogenetics
an approach that uses genetically-encoded constructs to precisely and non- invasively manipulate specific molecular process or cell function by light.
- Photoacoustic tomography (PAT)
an imaging technique that is based on photoacoustic effect and combines optical absorption and acoustic detection. The probe is excited with light but instead of fluorescence, thermoelastically generated acoustic waves are detected. Applied for deep tissue imaging, it allows to achieve higher spatial resolution at greater depths than purely optical imaging, because ultrasonic waves are scattered less than photons in tissue.
- Protein kinase A (PKA)
a cAMP-dependent serine/threonine kinase, a crucial enzyme in cell signaling cascades initiated by G protein coupled receptors (GPCRs) through cyclic AMP second messenger. It participates in several processes, including regulation of glycogen, sugar, lipid metabolism, and activation of reward system in the brain.
- Rac1 GTPase
a member of Rho family of small GTPases that regulate multiple signaling pathways controlling cell motility, proliferation, differentiation, and also glucose uptake. Since it is crucial for lamellipodia formation in cell motility, it is involved in cell invasion and metastasis in cancer.
- Structured illumination microscopy (SIM)
a widefield technique that relies on a grid pattern in one of the illumination apertures to extract information from the image focal plane and to produce an approximately two-fold increase in resolution in two or three dimensions.
- Two-photon (2P) microscopy
a microscopy technique based on two-photon excitation of fluorescence (two long-wavelength low-energy photons cause higher energy electronic transition in a fluorescent molecule. Confined focal volume and deeper penetrating NIR light result in cellular resolution up to 1 mm depth in tissue.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Shcherbakova DM et al. (2015) Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol 27, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y et al. (2013) In vivo imaging of orthotopic prostate cancer with far-red gene reporter fluorescence tomography and in vivo and ex vivo validation. J Biomed Opt 18 (10), 101305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiguet-Jiglaire C et al. (2014) Noninvasive near-infrared fluorescent protein-based imaging of tumor progression and metastases in deep organs and intraosseous tissues. J Biomed Opt 19 (1), 16019. [DOI] [PubMed] [Google Scholar]
- 4.Hock AK et al. (2014) iRFP is a sensitive marker for cell number and tumor growth in high- throughput systems. Cell Cycle 13 (2), 220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai CW et al. (2016) Using Dual Fluorescence Reporting Genes to Establish an In Vivo Imaging Model of Orthotopic Lung Adenocarcinoma in Mice. Mol Imaging Biol 18 (6), 849–859. [DOI] [PubMed] [Google Scholar]
- 6.Genevois C et al. (2016) In Vivo Follow-up of Brain Tumor Growth via Bioluminescence Imaging and Fluorescence Tomography. Int J Mol Sci 17 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao CN et al. (2018) Gene therapy for human glioblastoma using neurotropic JC virus-like particles as a gene delivery vector. Sci Rep 8 (1), 2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fyk-Kolodziej B et al. (2014) Marking cells with infrared fluorescent proteins to preserve photoresponsiveness in the retina. Biotechniques 57 (5), 245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richie CT et al. (2017) Near-infrared fluorescent protein iRFP713 as a reporter protein for optogenetic vectors, a transgenic Cre-reporter rat, and other neuronal studies. J Neurosci Methods 284, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piatkevich KD et al. (2017) Near-Infrared Fluorescent Proteins Engineered from Bacterial Phytochromes in Neuroimaging. Biophys J 113 (10), 2299–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y et al. (2014) Assessing in vitro stem-cell function and tracking engraftment of stem cells in ischaemic hearts by using novel iRFP gene labelling. J Cell Mol Med 18 (9), 1889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mezzanotte L et al. (2017) Optimized Longitudinal Monitoring of Stem Cell Grafts in Mouse Brain Using a Novel Bioluminescent/Near Infrared Fluorescent Fusion Reporter. Cell Transplant 26 (12), 1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bantounas I et al. (2018) Generation of Functioning Nephrons by Implanting Human Pluripotent Stem Cell-Derived Kidney Progenitors. Stem Cell Reports 10 (3), 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvo-Alvarez E et al. (2015) Infrared fluorescent imaging as a potent tool for in vitro, ex vivo and in vivo models of visceral leishmaniasis. PLoS Negl Trop Dis 9 (3), e0003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isomura M et al. (2017) Near-infrared fluorescent protein iRFP720 is optimal for in vivo fluorescence imaging of rabies virus infection. J Gen Virol 98 (11), 2689–2698. [DOI] [PubMed] [Google Scholar]
- 16.Filonov GS et al. (2011) Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol 29 (8), 757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shcherbakova DM and Verkhusha VV (2013) Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10 (8), 751–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shcherbakova DM et al. (2016) Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun 7, 12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shcherbakova DM et al. (2018) Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat Chem Biol 14 (6), 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu X et al. (2009) Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324 (5928), 804–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D et al. (2014) An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat Commun 5, 3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D et al. (2015) A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods 12 (8), 763–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auldridge ME et al. (2012) Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J Biol Chem 287 (10), 7000–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez EA et al. (2016) A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat Methods 13 (9), 763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shemetov AA et al. (2017) How to Increase Brightness of Near-Infrared Fluorescent Proteins in Mammalian Cells. Cell Chem Biol 24 (6), 758–766 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecoq J and Schnitzer MJ (2011) An infrared fluorescent protein for deeper imaging. Nat Biotechnol 29 (8), 715–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luker KE et al. (2015) Comparative study reveals better far-red fluorescent protein for whole body imaging. Sci Rep 5, 10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaner NC et al. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22 (12), 1567–72. [DOI] [PubMed] [Google Scholar]
- 29.Verkhusha VV et al. , Monomeric near-infrared fluorescent proteins engineered from bacterial phytochromes and methods for making same, U.S. Patent Application 15/497,667, 2017. [Google Scholar]
- 30.Donnelly SK et al. (2017) Rac3 regulates breast cancer invasion and metastasis by controlling adhesion and matrix degradation. J Cell Biol 216 (12), 4331–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssen AFJ et al. (2017) Myosin-V Induces Cargo Immobilization and Clustering at the Axon Initial Segment. Front Cell Neurosci 11, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinhard L et al. (2018) Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat Commun 9 (1), 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roman W et al. (2017) Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat Cell Biol 19 (10), 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishii T et al. (2015) Light generation of intracellular Ca(2+) signals by a genetically encoded protein BACCS. Nat Commun 6, 8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idevall-Hagren O et al. (2012) Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci U S A 109 (35), E2316–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zlobovskaya OA et al. (2016) Genetically encoded far-red fluorescent sensors for caspase-3 activity. Biotechniques 60 (2), 62–8. [DOI] [PubMed] [Google Scholar]
- 37.Shemiakina II et al. (2012) A monomeric red fluorescent protein with low cytotoxicity. Nat Commun 3, 1204. [DOI] [PubMed] [Google Scholar]
- 38.Bindels DS et al. (2017) mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14 (1), 53–56. [DOI] [PubMed] [Google Scholar]
- 39.Muller SM et al. (2013) Quantification of Forster resonance energy transfer by monitoring sensitized emission in living plant cells. Front Plant Sci 4, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galperin E et al. (2004) Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods 1 (3), 209–17. [DOI] [PubMed] [Google Scholar]
- 41.Subach OM et al. (2008) Conversion of red fluorescent protein into a bright blue probe. Chem Biol 15 (10), 1116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu N et al. (2011) Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell 22 (23), 4647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosco EE et al. (2009) Rac1 GTPase: a “Rac” of all trades. Cell Mol Life Sci 66 (3), 370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oeckinghaus A and Ghosh S (2009) The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1 (4), a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakaue-Sawano A et al. (2008) Visualizing spatiotemporal dynamics of multicellular cell- cycle progression. Cell 132 (3), 487–98. [DOI] [PubMed] [Google Scholar]
- 46.Tchekanda E et al. (2014) An infrared reporter to detect spatiotemporal dynamics of protein- protein interactions. Nat Methods 11 (6), 641–4. [DOI] [PubMed] [Google Scholar]
- 47.Filonov GS and Verkhusha VV (2013) A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol 20 (8), 1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey N et al. (2015) Combining random gene fission and rational gene fusion to discover near-infrared fluorescent protein fragments that report on protein-protein interactions. ACS Synth Biol 4 (5), 615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M et al. (2015) Novel near-infrared BiFC systems from a bacterial phytochrome for imaging protein interactions and drug evaluation under physiological conditions. Biomaterials 48, 97–107. [DOI] [PubMed] [Google Scholar]
- 50.Kodama Y and Hu CD (2010) An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques 49 (5), 793–805. [DOI] [PubMed] [Google Scholar]
- 51.To TL et al. (2015) Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci U S A 112 (11), 3338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stabley DR et al. (2015) Real-time fluorescence imaging with 20 nm axial resolution. Nat Commun 6, 8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svoboda K and Yasuda R (2006) Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron 50 (6), 823–39. [DOI] [PubMed] [Google Scholar]
- 54.Ji N et al. (2008) Advances in the speed and resolution of light microscopy. Curr Opin Neurobiol 18 (6), 605–16. [DOI] [PubMed] [Google Scholar]
- 55.Kobat D et al. (2011) In vivo two-photon microscopy to 1.6-mm depth in mouse cortex. J Biomed Opt 16 (10), 106014. [DOI] [PubMed] [Google Scholar]
- 56.Wang K et al. (2015) Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat Commun 6, 7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filonov GS et al. (2012) Deep-tissue photoacoustic tomography of a genetically encoded near-infrared fluorescent probe. Angew Chem Int Ed Engl 51 (6), 1448–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krumholz A et al. (2014) Multicontrast photoacoustic in vivo imaging using near-infrared fluorescent proteins. Sci Rep 4, 3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deliolanis NC et al. (2014) Deep-tissue reporter-gene imaging with fluorescence and optoacoustic tomography: a performance overview. Mol Imaging Biol 16 (5), 652–60. [DOI] [PubMed] [Google Scholar]
- 60.Yao J et al. (2016) Multiscale photoacoustic tomography using reversibly switchable bacterial phytochrome as a near-infrared photochromic probe. Nat Methods 13 (1), 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shcherbakova DM et al. (2015) Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins. Chem Biol 22 (11), 1540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baloban M et al. (2017) Designing brighter near-infrared fluorescent proteins: insights from structural and biochemical studies. Chem Sci 8 (6), 4546–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goedhart J et al. (2010) Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods 7 (2), 137–9. [DOI] [PubMed] [Google Scholar]
- 64.Repina NA et al. (2017) At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu Rev Chem Biomol Eng 8, 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rost BR et al. (2017) Optogenetic Tools for Subcellular Applications in Neuroscience. Neuron 96 (3), 572–603. [DOI] [PubMed] [Google Scholar]
- 66.Hochbaum DR et al. (2014) All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods 11 (8), 825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emiliani V et al. (2015) All-Optical Interrogation of Neural Circuits. J Neurosci 35 (41), 13917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Entcheva E and Bub G (2016) All-optical control of cardiac excitation: combined high- resolution optogenetic actuation and optical mapping. J Physiol 594 (9), 2503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barykina NV et al. (2016) A new design for a green calcium indicator with a smaller size and a reduced number of calcium-binding sites. Sci Rep 6, 34447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hock AK et al. (2017) Development of an inducible mouse model of iRFP713 to track recombinase activity and tumour development in vivo. Sci Rep 7 (1), 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tran MT et al. (2014) In vivo image analysis using iRFP transgenic mice. Exp Anim 63 (3), 311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin MZ et al. (2009) Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem. Biol. 16 (11), 1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wannier TM et al. (2017) Monomerization of Far-Red Fluorescent Proteins. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cranfill PJ et al. (2016) Quantitative assessment of fluorescent proteins. Nat Methods 13 (7), 557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rumyantsev KA et al. (2016) Near-infrared bioluminescent proteins for two-color multimodal imaging. Sci Rep 6, 36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rice WL et al. (2015) In vivo tomographic imaging of deep-seated cancer using fluorescence lifetime contrast. Cancer Res 75 (7), 1236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tzoumas S et al. (2015) Effects of multispectral excitation on the sensitivity of molecular optoacoustic imaging. J Biophotonics 8 (8), 629–37. [DOI] [PubMed] [Google Scholar]
- 78.Mark J et al. (2018) Dual-wavelength 3D photoacoustic imaging of mammalian cells using a photoswitchable phytochrome reporter protein. Communications Physics 1 (1), 3. [Google Scholar]
- 79.Telford WG et al. (2015) Multiparametric flow cytometry using near-infrared fluorescent proteins engineered from bacterial phytochromes. PLoS One 10 (3), e0122342. [DOI] [PMC free article] [PubMed] [Google Scholar]


