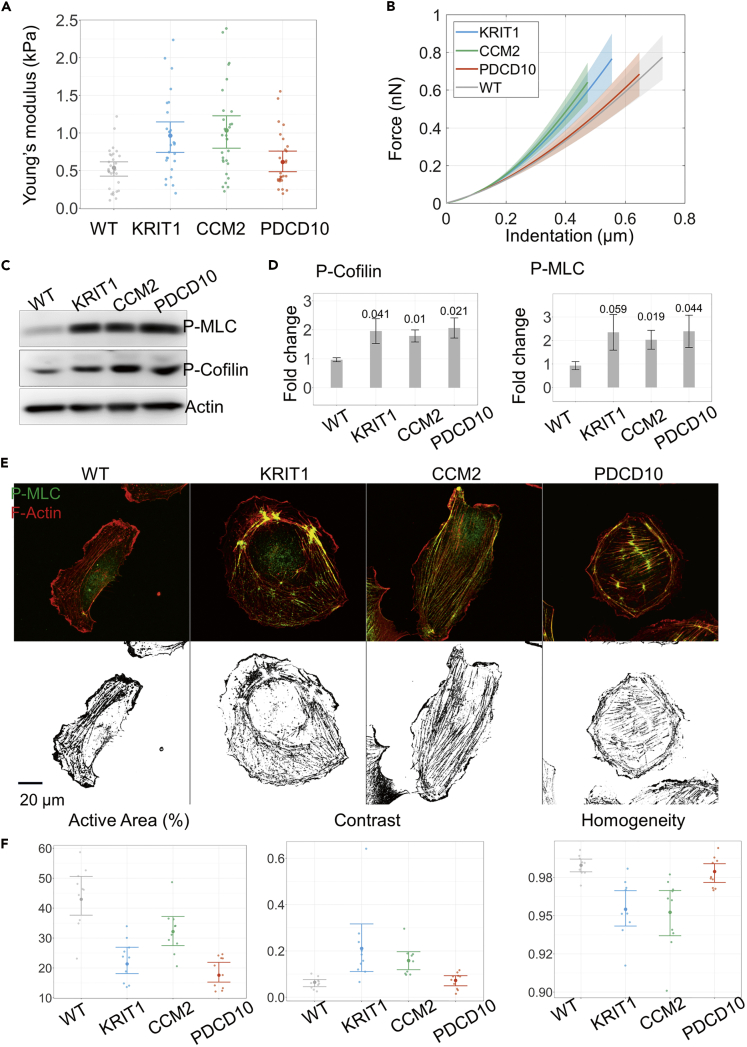
Figure 3.
Changes in Actomyosin Cytoskeleton Associated with Loss of CCM Protein Expression Result in Increased Stiffness of Cells with Knockdown of KRIT1 and CCM2 but Not PDCD10
(A) AFM stiffness measurement results for single PLKO.1 (WT) and cells with knockdown of KRIT1, CCM2, and PDCD10. The graph shows mean values and standard deviations of measurements from up to 30 cells in each group. Note higher stiffness of KRIT1 and CCM2 KD but not PDCD10 KD cells when compared with WT. The data shown are from one representative of three independent experiments.
(B) The graph shows representative force curves of WT cells and cells with loss of CCM protein expression recorded by AFM, with shaded area representing ±2·SEM.
(C) Representative immunoblot showing increased phosphorylation of myosin light chain (MLC-P) and cofilin in HUVEC cells with loss of KRIT1, CCM2, and PDCD10 expression. Total actin used as a loading control.
(D) Quantification of fold change of MLC-P and phospho-cofilin levels. Optical densities of the corresponding bands were normalized to the loading control; the results represent an increase over PLKO.1 (WT) expression levels (mean ± SEM, n = 3 experiments. In the bar graphs p values are indicated for each of the KD cultures compared with WT).
(E) Upper row: representative images of cells immunolabeled with an antibody against phosphorylated myosin light chain (MLC-P) (green), and incubated with rhodamine-phalloidin to visualize F-actin (red). Lower row: results of subtraction of thresholded signals from the F-actin and MLC-P channels from the images of cells above.
(F) Percentage of the active area defined here as a fraction of the pixels, which have an over-threshold intensity of the F-actin signal but a sub-threshold intensity of the MLC-P signal, among all pixels within a near-edge strip. Contrast and homogeneity are statistical measures of the image derived from the gray-level co-occurrence matrix (see Transparent Methods). Data are represented as mean ±2·SEM.