FIGURE 7.
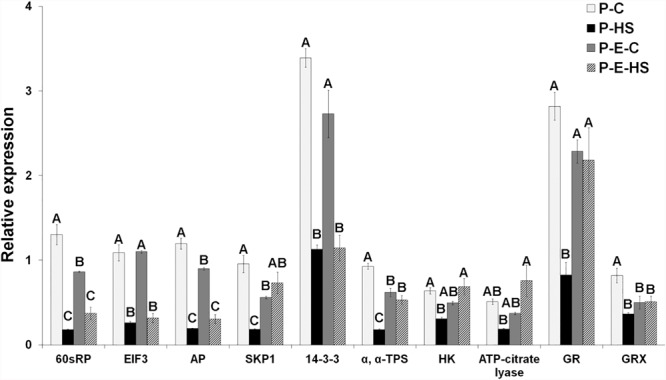
Tomato pollen transcript data for genes involved in protein synthesis and degradation, carbohydrate metabolism and signaling, TCA cycle, and redox regulation. The data was derived using qRT-PCR analyses and values were normalized relative to the expression levels of 18S rRNA in the same cDNA sample. qRT-PCR data are mean (±SE) of three biological replicates. For each gene, the bars with different letters are significantly different by multiple comparison Tukey’s HSD test (α = 0.05). Developing pollen grains were derived from: plants maintained at control conditions (P-C), plants exposed to HS conditions (P-HS), plants pre-treated with ethephon followed by maintaining the plants at control conditions (P-E-C), plants pre-treated with ethephon followed by exposing the plants to HS conditions (P-E-HS). AP, aspartic proteinase; EIF, eukaryotic initiation factor; GR, glutathione reductase; GRX, glutaredoxin; HK, hexokinase; RPs, ribosomal proteins; SKP1, s-phase kinase-associated protein 1; TPS, trehalose phosphate synthase. Corresponding Solyc numbers are given in Supplementary Table S13.
