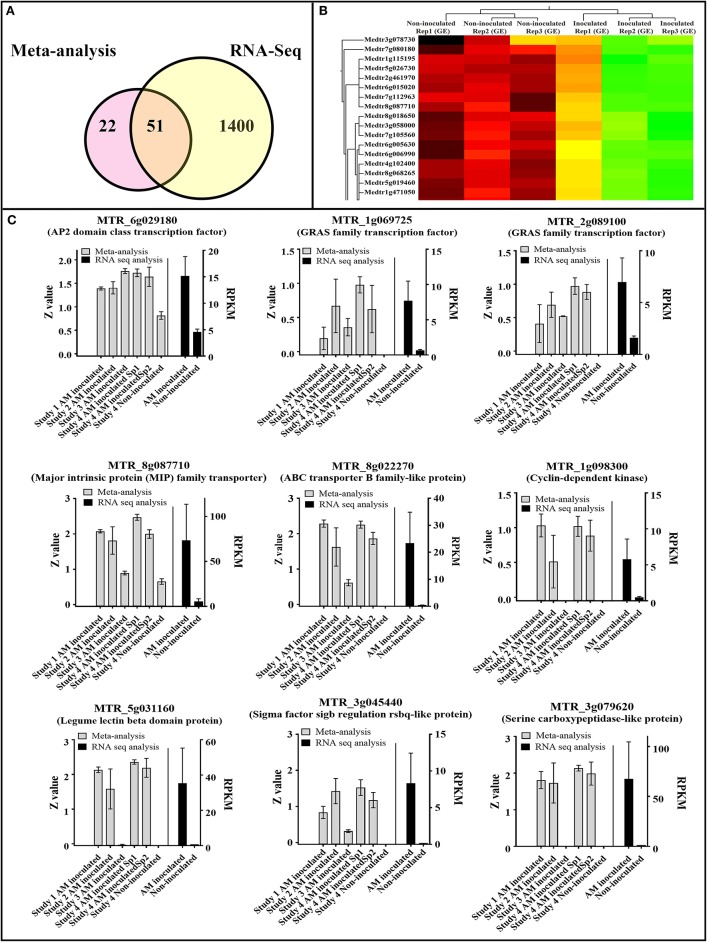
Figure 7.
High correspondence between RNA-seq data of Arbuscular mycorrhiza (AM) colonization and the identified AM colonization meta-signature in this study, derived from integration of meta-analysis with supervised attribute weighting models. (A) 51 out of 73 (70%) of the upregulated genes in the developed transcriptomic biosignature ofcolonization were also upregulated in the RNA-seq data of AM colonization with FDR-corrected p < 0.01. (B) The identified AM colonization meta-signature was able to accurately discriminate AM-inoculated samples from non-inoculated ones. (C) Visualization of the expression of some important genes of AM colonization signature in original experiments (based on standardized Z-value of expression) and RNA-seq experiment [based on RPKM (Reads Per Kilobase of transcript per Million mapped reads)].