Abstract
Near-infrared (NIR)-driven rhodopsins are of great interest in optogenetics and other optobiotechnological developments such as artificial photosynthesis and deep-tissue voltage imaging. Here we report that the proton pump proteorhodopsin (PR) containing a NIR-active retinal analogue (PR:MMAR) exhibits intense NIR fluorescence at a quantum yield of 3.3%. This is 130 times higher than native PR (Lenz M. O.; et al. Biophys J. 2006, 91, 255−262) and 3–8 times higher than the QuasAr and PROPS voltage sensors (Kralj J.; et al. Science 2011, 333, 345−348; Hochbaum D. R.; et al. Nat. Methods 2014, 11, 825–833). The NIR fluorescence strongly depends on the pH in the range of 6–8.5, suggesting potential application of MMAR-binding proteins as ultrasensitive NIR-driven pH and/or voltage sensors. Femtosecond transient absorption spectroscopy showed that upon near-IR excitation, PR:MMAR features an unusually long fluorescence lifetime of 310 ps and the absence of isomerized photoproducts, consistent with the high fluorescence quantum yield. Stimulated Raman analysis indicates that the NIR-absorbing species develops upon protonation of a conserved aspartate, which promotes charge delocalization and bond length leveling due to an additional methylamino group in MMAR, in essence providing a secondary protonated Schiff base. This results in much smaller bond length alteration along the conjugated backbone, thereby conferring significant single-bond character to the C13=C14 bond and structural deformation of the chromophore, which interferes with photoinduced isomerization and extends the lifetime for fluorescence. Hence, our studies allow for a molecular understanding of the relation between absorption/emission wavelength, isomerization, and fluorescence in PR:MMAR. As acidification enhances the resonance state, this explains the strong pH dependence of the NIR emission.
Rhodopsins are retinal-binding photoactivatable proteins1,2 with great impact as modular tools in optogenetics,2,3 voltage sensing,4−6 and artificial or engineered photosynthesis.7,8 A major challenge is designing microbial rhodopsins that can be activated by near-infrared light (NIR) for deep-tissue in vivo application or spectral window extension in oxygenic photosynthesis. There are several strategies for accessing red-shifted absorbance bands, e.g., by directed or random mutagenesis techniques,9 designing chimaeric proteins,10 and screening algal genomes.11 However, NIR-absorbing species that sufficiently maintain their desired activity have not yet been achieved with those approaches. Recently, NIR-driven proton-pumping and fluorescent rhodopsins were engineered using retinal analogues. Even though the retinal analogues need to be supplied to activate their function, in vivo applications have proved feasible.8,12,13
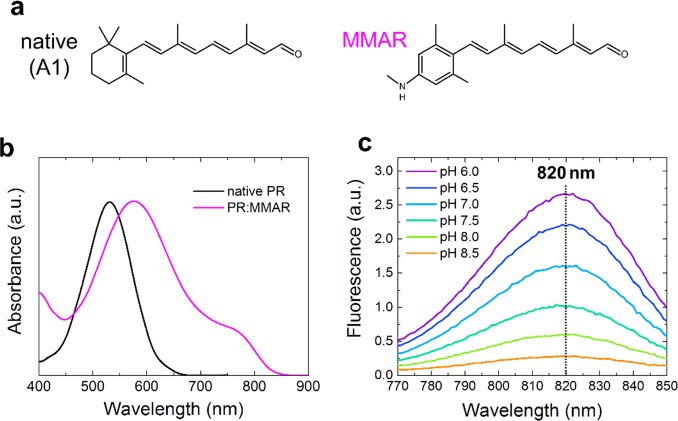
Native rhodopsins have a very low fluorescence quantum yield on the order of 10–4–10–5;14,15 in fact, it is 2.6 × 10–4 in proteorhodopsin (PR).16 The D97N mutant of PR (PR-D97N, also known as PROPS), which cannot sustain outward proton-pumping activity, has a relatively high fluorescence yield (ΦFL ≈ 0.01) and constitutes a suitable membrane voltage sensor in E. coli.4 Since then, various voltage sensors were designed based on microbial rhodopsins for visualization of action potentials of neurons, in particular, QuasArs5,6,13 and Archons.17 For an ideal pH or voltage sensor, a high fluorescence quantum yield and strong pH dependence of the emission intensity around physiological pH are required. Moreover, NIR absorption/emission will be beneficial for deeper tissue imaging where NIR has low absorbance and scattering.13,18 We previously reported that PR reconstituted with the retinal analogue 3-methylamino-16-nor-1,2,3,4-didehydroretinal (PR:MMAR, Figure 1a) shows far red-shifted absorbance and retains pump activity under illumination.12 Similar long-wavelength absorption bands were observed in bacteriorhodopsin reconstituted with azulenic or merocyanine dyes, but these proteins were photochemically inactive.19,20
Figure 1.
Chemical structure of MMAR and absorption/fluorescence spectra of PR:MMAR. (a) Structure of native retinal and its analogue MMAR. (b) Absorption spectra of native PR (PR:A1) and PR:MMAR at pH 7.0. (c) pH-dependent emission spectra of PR:MMAR at pH 6.0–8.5, at 760 nm excitation.
Figure 1b shows the PR:MMAR absorbance spectrum at pH 7.0, reproduced from ref (12), exhibiting a broad, heterogeneous absorption with bands and shoulders at around 570, 690, and 760 nm. The NIR absorption band near 760 nm has a low amplitude at this pH but increases dramatically upon acidification (Figure S1). We recorded the fluorescence properties of PR:MMAR to investigate potential application of MMAR-based microbial rhodopsins as a pH and/or voltage sensor. Figure 1c shows pH-dependent emission spectra of PR:MMAR at pH 6.0–8.5 with 760 nm excitation. Independent of the pH value, the emission peak was centered at 820 nm, which is about 120 nm red-shifted from native PR.16 The excitation spectrum of the 820 nm emission at pH 7.0 overlaps with the 1-transmittance spectrum as shown in Figure S2, clearly indicating that the NIR (>750 nm) absorbing form is the major emissive species. A fluorescence quantum yield of 3.3% was measured with 760 nm excitation at pH 7.0, which is 130-fold higher than that of native PR16 and 3–8 -fold higher than that of PROPS4 and QuasArs.5 Importantly, a strong pH dependence for the fluorescence intensity was observed over the pH range of 6.0–8.5 (Figure 1c), which clearly corresponds with the pH-dependent absorbance band near 760 nm (Figure S1). Considering this combination of properties, i.e., high fluorescence quantum yield, NIR absorption/emission, and strong pH dependence in the pH range of 6.0–8.5, we would advocate MMAR-binding retinal proteins as very promising candidates for ultrasensitive NIR pH and/or voltage sensors.
To elucidate the molecular origin of the intense fluorescence of PR:MMAR and its relation to the broad absorption profile, we applied broad-band femtosecond transient absorption spectroscopy up to the submillisecond time scale.21−24Figure 2a shows a kinetic trace at 750 nm upon excitation at 730 nm at pH 7.0 (blue symbols), where the NIR-absorbing state is selectively excited. Figure 2b shows the results of global analysis in terms of a sequential model (evolution-associated difference spectra, EADS, top panel) and a parallel, sum-of-exponentials model (decay-associated difference spectra, DADS, bottom panel). Kinetic traces and fits are presented in Figure S3. Three time constants are required to fit the data: 90 fs, 44 ps, and 310 ps. The gray EADS, which is created instantaneously after application of the pump pulse, has a negative signal from 620 to 720 nm that is assigned to ground-state bleach and a positive absorption at 580 nm that is assigned to excited-state absorption. This state evolves to the red EADS in 90 fs, which represents a further rise of the excited-state absorption band at 580 nm, with the ground state bleach at >650 nm remaining constant. The 90 fs time constant is assigned to ultrafast propagation out of the Franck–Condon region. The red EADS evolves to the blue EADS in 44 ps and involves only minor spectral changes. It is assigned to a minor population (<5%) that has an excited-state lifetime of 44 ps. Finally, the blue EADS has a lifetime of 310 ps and decays to zero amplitude.
Figure 2.
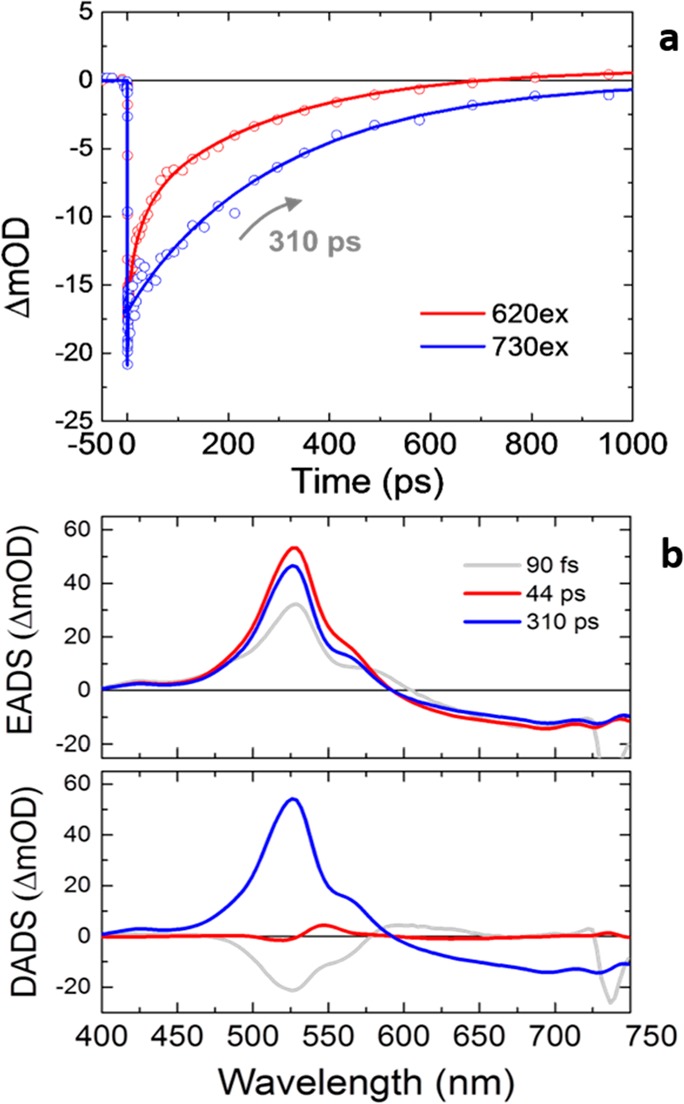
(a) Time traces of transient absorption experiments of PR:MMAR at pH 7 detected at 750 nm, upon 620 nm excitation (red circles) or 730 nm excitation (blue circles). The solid lines show fitting curves. (b) Evolution-associated difference spectra (EADS) (top) and decay-associated difference spectra (DADS) (bottom) upon excitation at 730 nm with fitted lifetimes indicated. The reader is referred to the Methods section for the interpretation of EADS and DADS.
We conclude that upon excitation at 730 nm, the excited state decays almost single-exponentially to zero in 310 ps. In addition, no photoproduct is detected, which implies that isomerization of the MMAR chromophore is largely suppressed. The excited-state lifetime (i.e., fluorescence lifetime) of 310 ps is unusually long for retinal proteins, which generally feature subpicosecond and picosecond lifetimes,1,16,21,25−30 and is consistent with the high fluorescence quantum yield.
The absorption spectrum of PR:MMAR is very broad, suggesting the presence of multiple spectral species (Figures 1b and S1). Upon excitation at shorter wavelengths, dynamics quite distinct from that at 730 nm excitation are observed. Upon 620 nm excitation, a multiexponential excited-state decay was observed with lifetimes of 6.4, 34, and 290 ps (Figure 2a, red symbols), along with formation of a photoproduct that resembles the well-known C13=C14 isomerized K-intermediate in microbial rhodopsins. Excitation at 510 and 670 nm gave similar results, albeit with different relative amplitudes of the lifetimes. A detailed account and global analysis of the excited-state dynamics at 620, 510, and 670 nm excitation wavelengths is presented in Figures S4–S6. Figure S7 shows that the 34 ps component is related to formation of the K-like isomerized photoproduct and that the 6.4 and 310 ps components represent nonproductive excited-state decay pathways to the initial ground state. In addition, in the cases that K was observed, the photocycle proceeded by formation of an M-like intermediate on submicrosecond and microsecond time scales, indicative of proton transfer from the MMAR Schiff base to Asp-97. We conclude that the broad absorption that spans the green to the NIR wavelengths is caused by three distinct electronic substates that have particular lifetimes and photochemical reactivity. Yet, the NIR absorption band near 760 nm is the only one presenting a single long-living excited-state component and significant fluorescence.
Proton-pumping activity in starved E. coli cells expressing PR:MMAR was observed at pH 7 with 617, 660, and 730 nm continuous LED illumination,12 which appears to be at odds with the present observation that for the NIR-absorbing species identified above the K- and M-like photoproducts were not detected. We first note that proton transfer from the protonated RSB may not be absolutely essential for proton-pumping in PR.31 Alternatively, PR:MMAR may behave slightly differently in DDM micelles (this study) as compared to the E. coli membrane environment (ref (12)), although no such differences were observed for native PR.32 It must be noted, however, that the proton-pumping rate of PR:MMAR is significantly smaller than that of native PR (PR:A1),12 indicating that this process is less robust in PR:MMAR and that the observed small difference between detergent-isolated PR:MMAR (no M-like state formation detected upon NIR excitation) and membrane-bound PR:MMAR (low proton-pumping activity under NIR illumination) may indeed relate to the microenvironment. It should also be noted that fluorescence and proton-pumping are not mutually exclusive for microbial rhodopsins.6
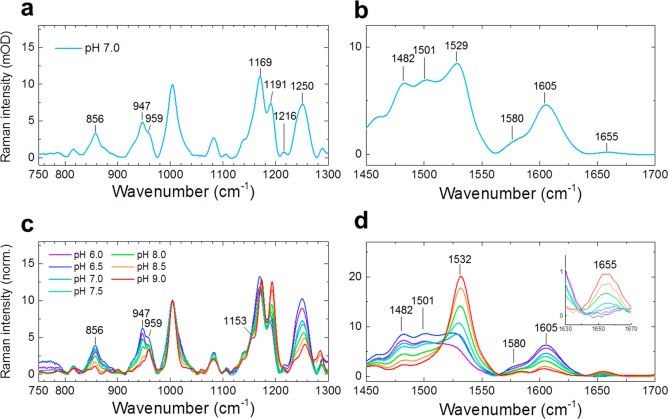
To investigate the ground-state configuration of the MMAR chromophore and its electronic structure, we applied preresonance watermarked stimulated Raman spectroscopy.33−35Figure 3a shows the ground-state stimulated Raman spectrum of PR:MMAR from 750 to 1300 cm–1 at pH 7.0. Various modes (1169, 1191, and 1216 cm–1) comply with C–C stretching modes in the native all-trans retinal chromophore in bacteriorhodopsin and PR.36,37 We furthermore identify isolated hydrogen-out-of-plane (HOOP) modes (856 cm–1), coupled HOOPs (947, 957 cm–1), and methyl rock (1004 cm–1).36 The 1250 cm–1 band is assigned to the aromatic amine C–N stretch that is particular to the MMAR chromophore.38Figure 3c shows stimulated Raman spectra over the same vibrational range between pH 6.0 and 9.0. The spectra were normalized at the 1004 cm–1 band. The ethylenic isolated (=C–H) and coupled (H–C=C–H) wags or HOOP vibrations are very sensitive to skeletal deformation: the strong increase of bands near 860 (isolated wags) and 950 cm–1 (coupled wags) upon lowering the pH is indicative of increasing distortion of the chromophore. The overall band distribution in the region of 1100–1220 cm–1 hardly varies over this pH range, except for a slight shift in frequencies, an increase in the intensity of the 1191 cm–1 mode (C14–C15 stretch39) with increasing pH, and concomitantly a more pronounced shoulder at ca. 1150 cm–1, possibly reflecting deprotonation of the Asp97 counterion.32 It should be noted that the C–C stretching modes are largely insensitive to electron delocalization,36,40 consistent with the very small frequency shifts at pH values between 6.0 and 9.0. The decrease of the 1191 cm–1 mode upon lowering the pH remains currently unexplained.
Figure 3.
pH-dependent ground-state stimulated Raman spectra of PR:MMAR. (a,b) Raman spectra at pH 7.0 at 750–1300 and 1450–1700 cm–1, respectively; (c,d) pH-dependent Raman spectra over the same vibrational ranges. The spectra were normalized at the 1004 cm–1 band.
In the C=C stretching region (∼1500–1600 cm–1) at pH 7.0 (Figure 3b,d), several Raman bands can be observed at 1482, 1501, 1528, 1580, and 1605 cm–1, which are strongly pH-dependent. Those at 1482, 1501, 1580, and 1605 cm–1 are very small at pH 8.5 and 9.0 and strongly increase upon acidification, while the single very strong band at 1532 cm–1 slightly downshifts to 1528 cm–1 and substantially decreases upon acidification. This pH dependence reflects that of the absorbance spectrum (Figure S1). Although some appear at an unusually low frequency, the bands between 1450 and 1550 cm–1 represent C=C stretching modes.36,41 This assignment stems from the observation that the C=C stretching frequency is correlated with the degree of electron delocalization in the chromophore and hence inversely correlated with the absorption maxima.42−44 Accordingly, the 1532 cm–1 mode would correspond to an absorbance band at around 560 nm, and the 1501 and 1482 cm–1 modes correspond to bands at around 690 and 750 nm, respectively, in excellent agreement with the absorbance spectra in Figure S1.
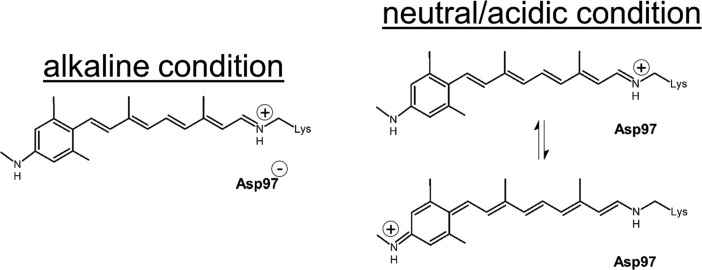
The 1655 cm–1 signal is assigned to the C=NH stretching mode, which is an indicator for the protonated retinal Schiff base widely seen in native rhodopsins.41,45 Remarkably, the 1655 cm–1 mode was replaced upon acidification by two peaks at ∼1580 and ∼1605 cm–1. If those peaks would represent a C=C stretching mode, we would expect a short-wavelength (blue-shifted below 400 nm) absorption species to appear at a lower pH, but this is not observed in the absorption spectra (Figure S1). Therefore, we do not assign the 1580 and 1605 cm–1 bands to C=C stretching modes but rather propose the formation of resonance species upon protonation of the Asp97 counterion, resulting in extensive electron delocalization involving both −N–C– elements (Figure 4). Because the pKa of the primary counterion (Asp97) varies between 6.5 and 7.7,12,46−49 this counterion is largely protonated at acidic pH, resulting in a weakened hydrogen bond between the retinal Schiff base and the counterion complex and less effective charge stabilization at the protonated retinal Schiff base. However, importantly, in MMAR, the charge can effectively delocalize toward the other methyl–N–C group on the aromatic ring at the other end of the chromophore, allowing a boundary structure with a second protonated Schiff base (Figure 4, right bottom), leading to a strong reduction in bond length alteration over the conjugated double bond system. This interpretation agrees with a lower C=NH stretch vibration frequency for both Schiff bases (Figure S8) as well as a strong increase of the bands at around 1250 cm–1 upon protonation (Figure S8), which could reflect an aromatic secondary amine stretch.38 Presumably, the two boundary structures shown in Figure 4 are in a pH-dependent equilibrium, resulting in a complex system of resonance states, yielding the heterogeneous absorbance and vibrational profiles of PR:MMAR under alkaline and acidic conditions. Such charge delocalization induced upon protonation will result in further hybridization of molecular orbitals, leading to the lower-energy broad absorbance bands near 690 and 760 nm.
Figure 4.
Boundary structures of MMAR in PR under alkaline and neutral/acidic conditions. The secondary site for Schiff base protonation is shown in the lower right panel.
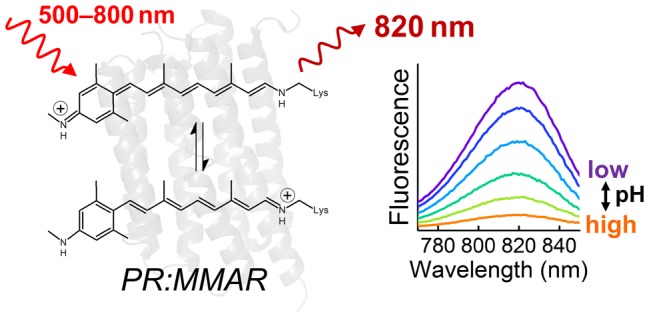
With the above results in hand, we are able to paint a molecular picture of the strong pH-dependent NIR fluorescence of PR:MMAR. Ultrafast spectroscopy revealed at least three distinct electronic substates. Upon NIR excitation at 730–760 nm, only the NIR fraction is populated and strong fluorescence results owing to the long excited-state lifetime of ∼300 ps, while under these conditions, no C13=C14 isomerization was detected. The stimulated Raman spectroscopy data confirm that such distinct electronic substates exist through the observation of three distinct C=C vibrational bands. Strikingly, upon acidification, a band rises that we attribute to the advent of resonance states that feature a boundary structure with another, secondary protonated Schiff base on the methyl–N–C group at the aromatic ring (Figure 4, bottom right). Notably, the NIR absorption and emission bands at 760 and 820 nm will originate from a state with a highly delocalized electron distribution, reminiscent of azulenic dyes50 and cyanine dyes.51,52 This will result in much smaller bond length alteration along the conjugated backbone, thereby conferring significant single-bond character to the C13=C14 bond and structural deformation of the chromophore, which interferes with photoinduced isomerization and extends the lifetime for fluorescence. This notion is crucial because in microbial rhodopsins, light-driven isomerization about the C13=C14 double bond is strongly catalyzed by the protein matrix and constitutes the principal excited-state deactivation channel. Thus, our study allows for a molecular understanding of the striking features of this promising retinal analogue protein system: strong pH-dependent NIR absorbance and fluorescence intensity and an excitation-wavelength-dependent photocycle. Additional molecular engineering of the protein and/or ligand may lead to further optimization of selected properties.
Acknowledgments
Y.H. and J.T.M.K. were supported by the Chemical Sciences Council of The Netherlands Organization for Scientific Research (NWO-CW) through a VICI grant to J.T.M.K. and a Middelgroot investment grant to J.T.M.K. S.G. and W.J.d.G. were supported by Leiden University and the research programme of BioSolar Cells (BSC Core Project Grant C2.9), cofinanced by the Dutch Ministry of Economic Affairs. This work was supported by the Czech Scientific Foundation Grant 17-01137S.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.8b02780.
Methods and pH-dependent absorption spectra, fluorescence excitation data, time-resolved data presented as kinetic traces, and global analysis results in the form of evolution-associated difference spectra (EADS), and decay-associated difference spectra (DADS) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ernst O. P.; Lodowski D. T.; Elstner M.; Hegemann P.; Brown L. S.; Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014, 114, 126–63. 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova E. G.; Sineshchekov O. A.; Li H.; Spudich J. L.. Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. In Annual Reviews in Biochemistry; Kornberg R. D., Ed.; 2017; Vol. 86, pp 845–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Vierock J.; Yizhar O.; Fenno L. E.; Tsunoda S.; Kianianmomeni A.; Prigge M.; Berndt A.; Cushman J.; Polle J.; Magnuson J.; Hegemann P.; Deisseroth K. The microbial opsin family of optogenetic tools. Cell 2011, 147, 1446–57. 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J. M.; Hochbaum D. R.; Douglass A. D.; Cohen A. E. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 2011, 333, 345–348. 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- Hochbaum D. R.; Zhao Y.; Farhi S. L.; Klapoetke N.; Werley C. A.; Kapoor V.; Zou P.; Kralj J. M.; Maclaurin D.; Smedemark-Margulies N.; Saulnier J. L.; Boulting G. L.; Straub C.; Cho Y. K.; Melkonian M.; Wong G. K. S.; Harrison D. J.; Murthy V. N.; Sabatini B. L.; Boyden E. S.; Campbell R. E.; Cohen A. E. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 2014, 11, 825–833. 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J. M.; Douglass A. D.; Hochbaum D. R.; Maclaurin D.; Cohen A. E. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 2012, 9, 90–U130. 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassens N. J.; Sousa D. Z.; dos Santos V.; de Vos W. M.; van der Oost J. Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol. 2016, 14, 692–706. 10.1038/nrmicro.2016.130. [DOI] [PubMed] [Google Scholar]
- Chen Q.; van der Steen J. B.; Arents J. C.; Hartog A. F.; Ganapathy S.; de Grip W. J.; Hellingwerf K. J. Deletion of sll1541 in Synechocystis sp strain PCC 6803 allows formation of a far-red-shifted holo-proteorhodopsin in vivo. Appl. Environ. Microbiol. 2018, 10.1128/AEM.02435-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L.; Yizhar O.; Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011, 34, 389–412. 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbguth K.; Prigge M.; Schneider F.; Hegemann P.; Gottschalk A. Bimodal activation of different neuron classes with the spectrally red-shifted channelrhodopsin chimera C1V1 in Caenorhabditis elegans. PLoS One 2012, 7, e46827. 10.1371/journal.pone.0046827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke N. C.; Murata Y.; Kim S. S.; Pulver S. R.; Birdsey-Benson A.; Cho Y. K.; Morimoto T. K.; Chuong A. S.; Carpenter E. J.; Tian Z.; Wang J.; Xie Y.; Yan Z.; Zhang Y.; Chow B. Y.; Surek B.; Melkonian M.; Jayaraman V.; Constantine-Paton M.; Wong G. K.; Boyden E. S. Independent optical excitation of distinct neural populations. Nat. Methods 2014, 11, 338–46. 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy S.; Venselaar H.; Chen Q.; de Groot H. J.; Hellingwerf K. J.; de Grip W. J. Retinal-Based Proton Pumping in the Near Infrared. J. Am. Chem. Soc. 2017, 139, 2338–2344. 10.1021/jacs.6b11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwig L.; Rice A. J.; Bedbrook C. N.; Zhang R. J. K.; Lignell A.; Cahn J. K. B.; Renata H.; Dodani S. C.; Cho I.; Cai L.; Gradinaru V.; Arnold F. H. Directed evolution of a bright near-infrared fluorescent rhodopsin using a synthetic chromophore. Cell Chem. Biol. 2017, 24, 415–425. 10.1016/j.chembiol.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindjee R.; Becher B.; Ebrey T. G. Fluorescence from chromophore of purple membrane-protein. Biophys. J. 1978, 22, 67–77. 10.1016/S0006-3495(78)85471-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochendoerfer G. G.; Mathies R. A. Spontaneous emission study of the femtosecond isomerization dynamics of rhodopsin. J. Phys. Chem. 1996, 100, 14526–14532. 10.1021/jp960509+. [DOI] [Google Scholar]
- Lenz M. O.; Huber R.; Schmidt B.; Gilch P.; Kalmbach R.; Engelhard M.; Wachtveitl J. First steps of retinal photoisomerization in proteorhodopsin. Biophys. J. 2006, 91, 255–62. 10.1529/biophysj.105.074690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich K. D.; Jung E. E.; Straub C.; Linghu C. Y.; Park D.; Suk H. J.; Hochbaum D. R.; Goodwin D.; Pnevmatikakis E.; Pak N.; Kawashima T.; Yang C. T.; Rhoades J. L.; Shemesh O.; Asano S.; Yoon Y. G.; Freifeld L.; Saulnier J. L.; Riegler C.; Engert F.; Hughes T.; Drobizhev M.; Szabo B.; Ahrens M. B.; Flavell S. W.; Sabatini B. L.; Boyden E. S. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 2018, 14, 352. 10.1038/s41589-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova D. M.; Baloban M.; Verkhusha V. V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr. Opin. Chem. Biol. 2015, 27, 52–63. 10.1016/j.cbpa.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner W.; Buss V.; Martin H. D.; Hoischen D.; Steinmuller S. Merocyanines as extremely bathochromically absorbing chromophores in the halobacterial membrane protein bacteriorhodopsin. Angew. Chem., Int. Ed. Engl. 1997, 36, 1630–1633. 10.1002/anie.199716301. [DOI] [Google Scholar]
- Asato A. E.; Li X. Y.; Mead D.; Patterson G. M. L.; Liu R. S. H. Azulenic retinoids and the corresponding bacteriorhodopsin analogs - unusually red-shifted pigments. J. Am. Chem. Soc. 1990, 112, 7398–7399. 10.1021/ja00176a051. [DOI] [Google Scholar]
- Hontani Y.; Marazzi M.; Stehfest K.; Mathes T.; van Stokkum I. H. M.; Elstner M.; Hegemann P.; Kennis J. T. M. Reaction dynamics of the chimeric channelrhodopsin C1C2. Sci. Rep. 2017, 7, 7217. 10.1038/s41598-017-07363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berera R.; van Grondelle R.; Kennis J. T. M. Ultrafast transient absorption spectroscopy: principles and application to photosynthetic systems. Photosynth. Res. 2009, 101, 105–118. 10.1007/s11120-009-9454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes T.; Heilmann M.; Pandit A.; Zhu J. Y.; Ravensbergen J.; Kloz M.; Fu Y. A.; Smith B. O.; Christie J. M.; Jenkins G. I.; Kennis J. T. M. Proton-coupled electron transfer constitutes the photoactivation mechanism of the plant photoreceptor UVR8. J. Am. Chem. Soc. 2015, 137, 8113–8120. 10.1021/jacs.5b01177. [DOI] [PubMed] [Google Scholar]
- Ravensbergen J.; Abdi F. F.; van Santen J. H.; Frese R. N.; Dam B.; van de Krol R.; Kennis J. T. M. Unraveling the carrier dynamics of BiVO4: a femtosecond to microsecond transient absorption study. J. Phys. Chem. C 2014, 118, 27793–27800. 10.1021/jp509930s. [DOI] [Google Scholar]
- Mathies R. A.; Brito Cruz C. H.; Pollard W. T.; Shank C. V. Direct observation of the femtosecond excited-state cis-trans isomerization in bacteriorhodopsin. Science 1988, 240, 777–779. 10.1126/science.3363359. [DOI] [PubMed] [Google Scholar]
- Rupenyan A.; van Stokkum I. H.; Arents J. C.; van Grondelle R.; Hellingwerf K. J.; Groot M. L. Reaction pathways of photoexcited retinal in proteorhodopsin studied by pump-dump-probe spectroscopy. J. Phys. Chem. B 2009, 113, 16251–6. 10.1021/jp9065289. [DOI] [PubMed] [Google Scholar]
- Tahara S.; Takeuchi S.; Abe-Yoshizumi R.; Inoue K.; Ohtani H.; Kandori H.; Tahara T. Ultrafast photoreaction dynamics of a light-driven sodium-ion-pumping retinal protein from Krokinobacter eikastus revealed by femtosecond time-resolved absorption spectroscopy. J. Phys. Chem. Lett. 2015, 6, 4481–4486. 10.1021/acs.jpclett.5b01994. [DOI] [PubMed] [Google Scholar]
- Hontani Y.; Broser M.; Silapetere A.; Krause B. S.; Hegemann P.; Kennis J. T. M. The femtosecond-to-second photochemistry of red-shifted fast-closing anion channelrhodopsin PsACR1. Phys. Chem. Chem. Phys. 2017, 19, 30402–30409. 10.1039/C7CP06414D. [DOI] [PubMed] [Google Scholar]
- Verhoefen M. K.; Bamann C.; Blocher R.; Forster U.; Bamberg E.; Wachtveitl J. The photocycle of channelrhodopsin-2: ultrafast reaction dynamics and subsequent reaction steps. ChemPhysChem 2010, 11, 3113–22. 10.1002/cphc.201000181. [DOI] [PubMed] [Google Scholar]
- Schnedermann C.; Muders V.; Ehrenberg D.; Schlesinger R.; Kukura P.; Heberle J. Vibronic dynamics of the ultrafast all-trans to 13-cis photoisomerization of retinal in Channelrhodopsin-1. J. Am. Chem. Soc. 2016, 138, 4757–4762. 10.1021/jacs.5b12251. [DOI] [PubMed] [Google Scholar]
- Xiao Y. W.; Partha R.; Krebs R.; Braiman M. Time-resolved FTIR spectroscopy of the photointermediates involved in fast transient H+ release by proteorhodopsin. J. Phys. Chem. B 2005, 109, 634–641. 10.1021/jp046314g. [DOI] [PubMed] [Google Scholar]
- Dioumaev A. K.; Brown L. S.; Shih J.; Spudich E. N.; Spudich J. L.; Lanyi J. K. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry 2002, 41, 5348–58. 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- Hontani Y.; Inoue K.; Kloz M.; Kato Y.; Kandori H.; Kennis J. T. The photochemistry of sodium ion pump rhodopsin observed by watermarked femto- to submillisecond stimulated Raman spectroscopy. Phys. Chem. Chem. Phys. 2016, 18, 24729–36. 10.1039/C6CP05240A. [DOI] [PubMed] [Google Scholar]
- Kloz M.; Weissenborn J.; Polivka T.; Frank H. A.; Kennis J. T. M. Spectral watermarking in femtosecond stimulated Raman spectroscopy: resolving the nature of the carotenoid S-star state. Phys. Chem. Chem. Phys. 2016, 18, 14619–14628. 10.1039/C6CP01464J. [DOI] [PubMed] [Google Scholar]
- Hontani Y.; Kloz M.; Polivka T.; Shukla M. K.; Sobotka R.; Kennis J. T. M. Molecular origin of photoprotection in cyanobacteria probed by watermarked femtosecond stimulated Raman spectroscopy. J. Phys. Chem. Lett. 2018, 9, 1788–1792. 10.1021/acs.jpclett.8b00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O.; Pardoen J. A.; Lugtenburg J.; Mathies R. A. Vibrational analysis of the 13-cis-retinal chromophore in dark-adapted bacteriorhodopsin. J. Phys. Chem. 1987, 91, 804–819. 10.1021/j100288a011. [DOI] [Google Scholar]
- Krebs R. A.; Dunmire D.; Partha R.; Braiman M. S. Resonance Raman characterization of proteorhodopsin’s chromophore environment. J. Phys. Chem. B 2003, 107, 7877–7883. 10.1021/jp034574c. [DOI] [Google Scholar]
- Socrates G.Infrared and Raman Characteristic Group Frequencies: tables and charts, 3rd ed.; John Wiley and Sons: Chichester, U.K., 2001. [Google Scholar]
- Smith S. O.; Braiman M. S.; Myers A. B.; Pardoen J. A.; Courtin J. M. L.; Winkel C.; Lugtenburg J.; Mathies R. A. Vibrational analysis of the all-trans-retinal chromophore in light-adapted bacteriorhodopsin. J. Am. Chem. Soc. 1987, 109, 3108–3125. 10.1021/ja00244a038. [DOI] [Google Scholar]
- Stockburger M.; Alshuth T.; Oesterhelt D.; Gärtner W.. Resonance Raman spectroscopy of bacteriorhodopsin: structure and function. In Spectroscopy of Biological Systems, Clark R. J. H., Hester R. E., Eds.; John Wiley and Sons: New York, 1986; pp 483–535. [Google Scholar]
- Smith S. O.; Braiman M. S.; Myers A. B.; Pardoen J. A.; Courtin J. M. L.; Winkel C.; Lugtenburg J.; Mathies R. A. Vibrational analysis of the all-trans-retinal chromophore in light-adapted bacteriorhodopsin. J. Am. Chem. Soc. 1987, 109, 3108–3125. 10.1021/ja00244a038. [DOI] [Google Scholar]
- Doukas A. G.; Aton B.; Callender R. H.; Ebrey T. G. Resonance Raman studies of bovine metarhodopsin I and metarhodopsin II. Biochemistry 1978, 17, 2430–5. 10.1021/bi00605a028. [DOI] [PubMed] [Google Scholar]
- Kajimoto K.; Kikukawa T.; Nakashima H.; Yamaryo H.; Saito Y.; Fujisawa T.; Demura M.; Unno M. Transient resonance raman spectroscopy of a light-driven sodium-ion-pump rhodopsin from Indibacter alkaliphilus. J. Phys. Chem. B 2017, 121, 4431–4437. 10.1021/acs.jpcb.7b02421. [DOI] [PubMed] [Google Scholar]
- Ogren J. I.; Mamaev S.; Russano D.; Li H.; Spudich J. L.; Rothschild K. J. Retinal chromophore structure and Schiff base interactions in red-shifted Channelrhodopsin-1 from Chlamydomonas augustae. Biochemistry 2014, 53, 3961–3970. 10.1021/bi500445c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun S.; Stoeppler D.; Keidel A.; Kuhlmann U.; Luck M.; Diehl A.; Geiger M. A.; Woodmansee D.; Trauner D.; Hegemann P.; Oschkinat H.; Hildebrandt P.; Stehfest K. Light-dark adaptation of channelrhodopsin involves photoconversion between the all-trans and 13-cis retinal isomers. Biochemistry 2015, 54, 5389–400. 10.1021/acs.biochem.5b00597. [DOI] [PubMed] [Google Scholar]
- Friedrich T.; Geibel S.; Kalmbach R.; Chizhov I.; Ataka K.; Heberle J.; Engelhard M.; Bamberg E. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol. 2002, 321, 821–838. 10.1016/S0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- Varo G.; Brown L. S.; Lakatos M.; Lanyi J. K. Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 2003, 84, 1202–7. 10.1016/S0006-3495(03)74934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S.; Kinnebrew M.; Schonenbach N. S.; Aye E.; Han S. Functional consequences of the oligomeric assembly of proteorhodopsin. J. Mol. Biol. 2015, 427, 1278–1290. 10.1016/j.jmb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T.; Weber I.; Glaubitz C.; Wachtveitl J. Proteorhodopsin photocycle kinetics between pH 5 and pH 9. Photochem. Photobiol. 2017, 93, 762–771. 10.1111/php.12753. [DOI] [PubMed] [Google Scholar]
- Muthyala R.; Watanabe D.; Asato A. E.; Liu R. S. H. The nature of the delocalized cations in azulenic bacteriorhodopsin analogs. Photochem. Photobiol. 2001, 74, 837–845. 10.1562/0031-8655(2001)0740837TNOTDC2.0.CO2. [DOI] [PubMed] [Google Scholar]
- Escobedo J. O.; Rusin O.; Lim S.; Strongin R. M. NIR dyes for bioimaging applications. Curr. Opin. Chem. Biol. 2010, 14, 64–70. 10.1016/j.cbpa.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitus M.; Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q. Rev. Biophys. 2011, 44, 123–151. 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.