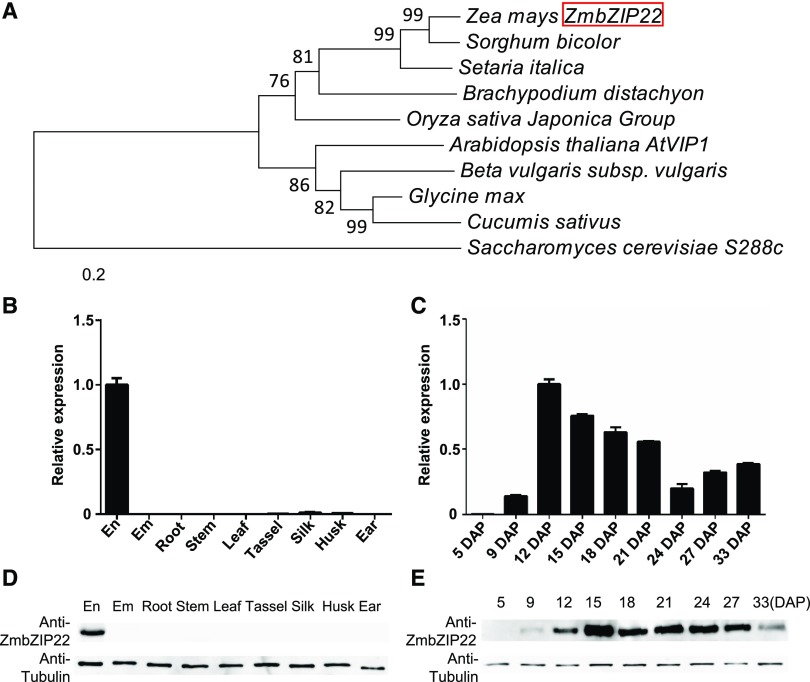
Figure 2.
Phylogenetic Relationships and Expression Pattern of ZmbZIP22.
(A) Phylogenetic relationships of ZmbZIP22 and its homologs in plants. Distances were estimated using the neighbor-joining algorithm. The numbers at the nodes (100) represent the percentage of 1000 bootstraps. The scale bar indicates the average number of amino acid substitutions per site. The S. cerevisiae homologous protein was used as an outgroup.
(B) and (C) Expression analysis of ZmbZIP22 by RT-qPCR. RT-qPCR analysis of ZmbZIP22 expression in various tissues (B) and developing kernels (C). The endosperm (En) and embryo (Em) samples in (B) were harvested at 15 DAP. Other tissues (root, stem, third leaf, tassel, silk, husk, and ear) were collected from field-cultivated W22 plants at the V12 stage. The kernel samples in (C) were collected at different developmental stages, as indicated by days after pollination. The Ubiquitin gene was used as internal control. Three biological replicates were made with tissues from three plants. Error bars indicate ±sd (n = 3).
(D) and (E) Immunoblot analysis of ZmbZIP22 expression in various tissues (D) and developing kernels (E). The embryo and endosperm samples in (D) were harvested at 15 DAP. Other tissues (root, stem, third leaf, tassel, silk, husk, and ear) were collected from field-cultivated W22 plants at the V12 stage. The kernel samples were collected at different developmental stages, as indicated by days after pollination. Antitubulin was used as a loading control.