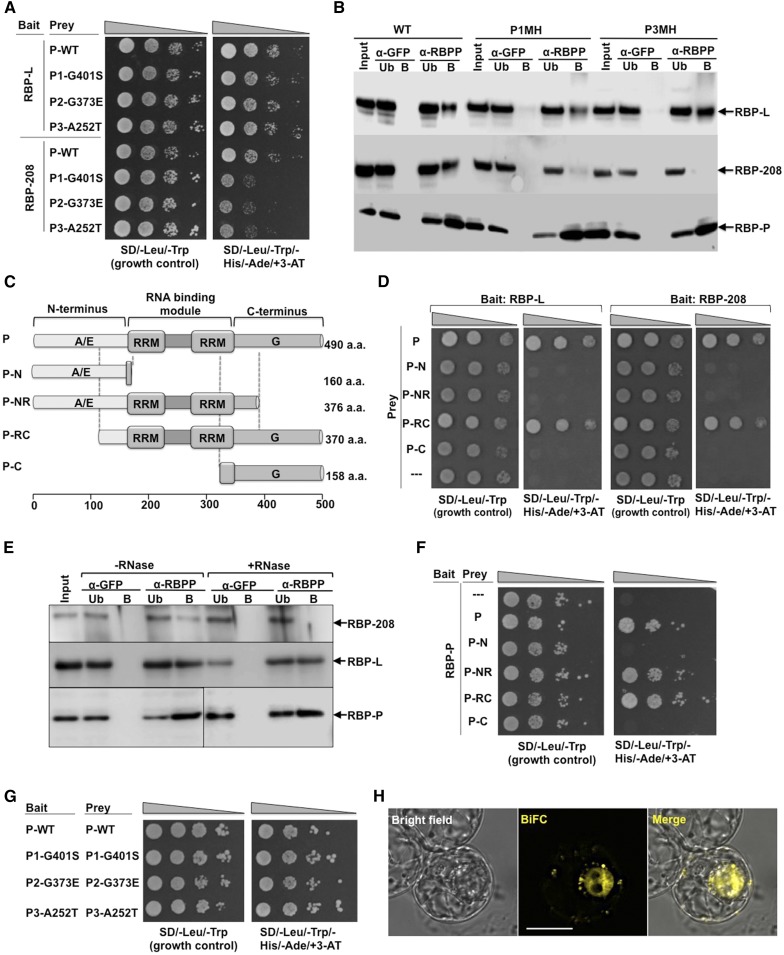
Figure 8.
Protein-Protein Interactions of RBP-P with Itself, RBP-L, and RBP-208.
(A) and (B) The effect of mutations (G401S, G373E, and A252T) on RBP-P interactions with RBP-L and RBP-208 as revealed by Y2H (A) and co-IP analyses (B) compared with the wild type. Note the retardation of growth of yeast clones carrying RBP-208 and mutant RBP-Ps (A) and the absence of RBP-208 in anti-RBP IPs (B). Input, starting rice grain lysate; Ub, unbound fraction; B, elution of the bound fraction from IP; α-GFP and α-RBPP, samples from anti-GFP and anti-RBP-P columns, respectively. Arrows indicate the presence of RBP-L, RBP-208, and RBP-P in various immunoprecipitates formed by anti-RBP-P but not by anti-GFP.
(C) Schematic structure of truncated RBP-P protein. aa, amino acids.
(D) Interactions of RBP-L (left panel) and RBP-208 (right panel) with intact and truncated RBP-P as viewed by Y2H analyses.
(E) Co-IP using anti-RBP-P antibody to detect its interaction with RBP-L and RBP-208 in developing rice grain extracts pretreated with or without RNase. Arrows denote RBP-P, RBP-L, and RBP-208 in immunoblot analyses.
(F) Identification of binding sites responsible for homodimerization of RBP-P. Note that dimer formation requires the two RRM domains, as interaction is seen with P-NR and P-RC but not with P-N or P-C.
(G) Dimerization analysis of wild-type and mutant RBP-P forms viewed by Y2H.
(H) BiFC analysis reveals the in vivo dimerization of RBP-P. nEYFP-RBP-P complemented with cEYFP-RBP-P for coexpression in BY-2 protoplasts. Bar = 20 μm.