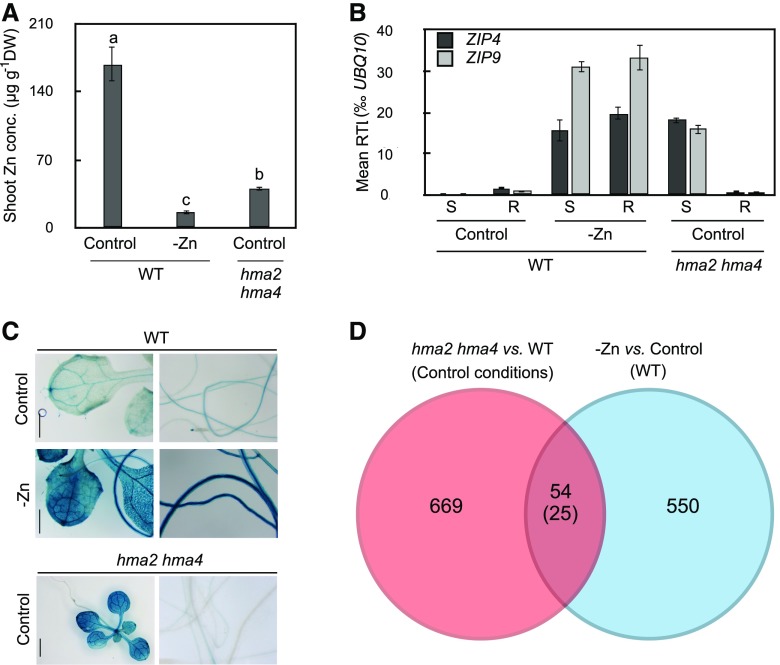
Figure 1.
Experimental Design and Validation, and Results of Transcriptomics Targeting Systemic Zn Deficiency Response of Roots.
(A) Shoot Zn concentrations in 21-d-old wild-type seedlings grown on Zn-deficient medium (−Zn) and hma2 hma4 plants grown on control agar-solidified medium for 14 d, subsequent to an initial cultivation on agar-solidified 0.5× MS medium for 7 d.
(B) ZIP4 and ZIP9 relative transcript levels in shoot (S) and root (R) of WT and hma2 hma4 seedlings, based on RT-qPCR. Seedlings were grown as in (A).
(C) GUS activity in ProZIP4:GUS wild-type and hma2 hma4 seedlings. Pictures are representative of 12 independent transgenic lines in Col-0 background, of which one representative line was used to introduce ProZIP4:GUS into the hma2 hma4 background by crossing. Shown are F3 seedlings that are homozygous for the ProZIP4:GUS transgene and for the hma2 and hma4 mutations. Seedlings were grown as in (A). Bars = 5 mm (WT) and 10 mm (hma2 hma4).
(D) Summary of microarray data. Shown are the number of genes showing ≥2-fold differences in transcript levels (in parentheses for same direction of changes in both comparisons).
Bar graphs show arithmetic mean ± sd (n = 3 pools of 16 plants, with each pool from one plate [A]; n = 4 technical replicates using tissue pooled from three plates, each with 16–18 seedlings [B]). Distinct letters indicate significant differences (P < 0.05) according to a Bonferroni-corrected Student’s t test.