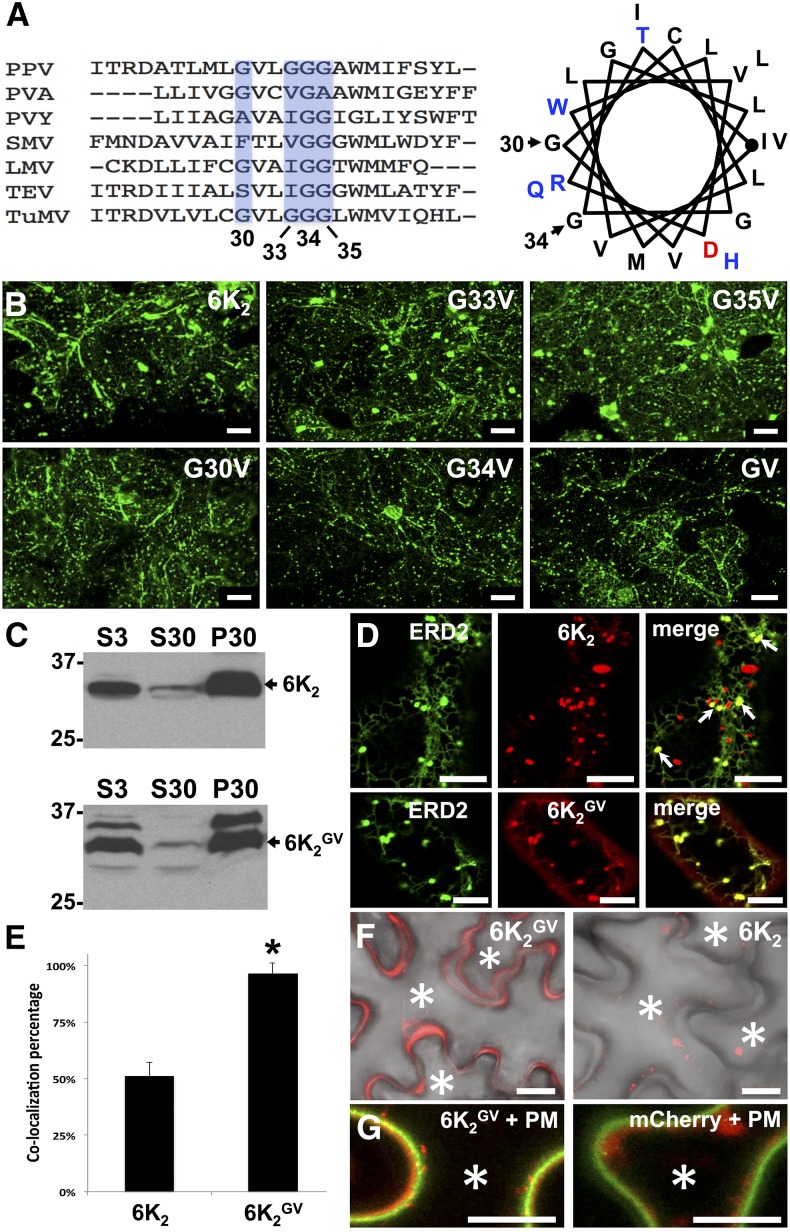
Figure 1.
Mutation of the GxxxG Motif in 6K2 Alters Its Localization.
(A) Sequence alignment of the predicted TMDs of several potyviral 6K2 proteins. The conserved Gly residues are shown (light blue) and the corresponding amino acid position in TuMV is indicated below. PPV, Plum pox virus; PVA, Potato virus A; PVY, Potato virus Y; SMV, Soybean mosaic virus; LMV, Lettuce mosaic virus; TEV, Tobacco etch virus. To the right is a helical wheel projection of the predicted 6K2 TMD of TuMV. The Gly residues located at positions 30 and 34 are indicated.
(B) Fluorescence imaging of N. benthamiana cells expressing GFP fusions of wild-type 6K2 and the indicated 6K2 mutants at 3 dpi. These images are three-dimensional renderings stacks of 40 1-μm-thick slices that overlap by 0.5 μm.
(C) Membrane association of wild-type 6K2 and 6K2GV protein. S3, total protein fraction; S30, soluble protein fraction; P30, membrane protein fraction. Immunoblotting was performed with anti-GFP antibodies.
(D) Confocal images of N. benthamiana epidermal cells expressing 6K2:mCherry (top middle panel) or 6K2GV:mCherry (bottom middle panel) with the cis-Golgi marker ERD2:GFP (left panels). Their coexpression is shown in the right panels (merge). Arrows in right top panel indicate 6K2:mCherry colocalizing with the cis Golgi marker.
(E) Colocalization of ERD2:GFP and 6K2:mCherry (6K2) or 6K2GV:mCherry (6K2GV). Statistical significance was calculated using the Pearson’s correlation coefficient r values of ImageJ and a t test analysis was done (*P < 0.05). Values indicate means ± sd and n = 18 cells.
(F) Fluorescence images of cells expressing 6K2GV:mCherry or 6K2:mCherry merged with the bright field image are shown.
(G) The PM marker AHA2:GFP was expressed with either 6K2GV:mCherry or mCherry. Asterisks indicate the interior of the cells. All confocal images are single optical images (1 μm thick). Three independent biological repeats were performed for (B), (D), and (F), and two for (C) and (G). Bars = 20 μm in (B), (F), and (G) and 10 μm in (D).