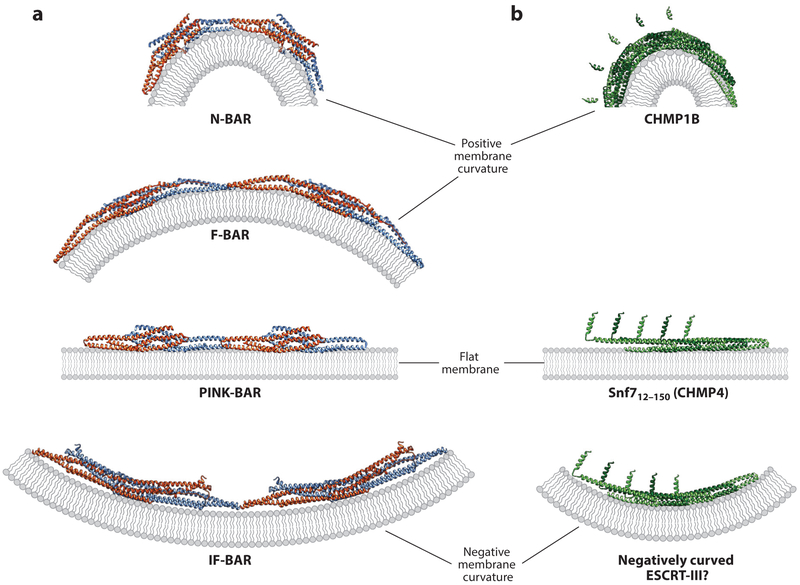
Figure 4. Models for stabilization of curved and flat membranes by BAR domain and ESCRT-III proteins.
(a) Illustrations showing how changing the angle between the end-associated BAR domain dimers (blue and orange subunits) can stabilize a continuum of differentially curved membranes (gray). Pairs of dimers from continuous BAR domain assemblies (end-on views) are shown in each case. Structural models are based on Mim & Unger (2012) and Mim et al. (2012) for the N-BAR case, Shimada et al. (2007) and Frost et al. (2008) for the F-BAR case, Guerrier et al. (2009) and Sporny et al. (2017) for the IF-BAR case, and Pykalainen et al. (2011) for the PINK-BAR case, (b) Illustrations showing how changes in intrinsic filament curvature could similarly allow ESCRT-III filaments (green) to stabilize a continuum of differentially curved membranes. (Top to bottom) A CHMP1B strand from the IST1NTD/CHMP1B filament bound to a stylized membrane (from McCullough et al. 2015), structure of the linear strand of Snf712–150 (CHMP4) from a crystal lattice (from Tang et al. 2015) bound to a stylized membrane, and a hypothetical strand of Snf712–150 (CHMP4) subunits bound to a negatively curved membrane. The Snf712—150 (CHMP4) strand in the bottom panel was modeled by altering the angle between each successive subunit in the strand shown in the middle panel.