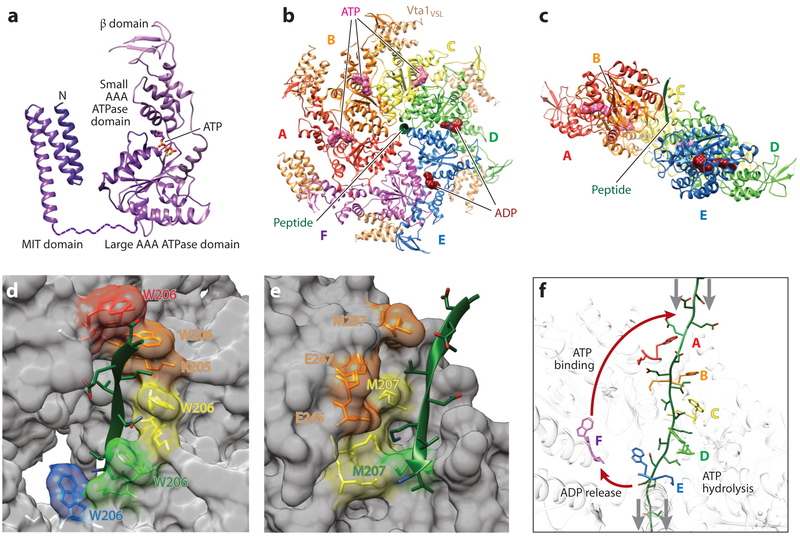
Figure 5. Structure, assembly, and mechanism of the Vps4 ATPase.
(a) Domain structure of a Vps4 monomer in complex with ATP. Structures are reproduced from Han et al. (2017) (ATPase cassette) and Obita et al. (2007) (MIT domain), (b) Top view of the asymmetric yeast Vps4 ring hexamer (subunits A–F) with associated dimeric VSL domains from Vta1 (beige), ESCRT-III peptide (green), and nucleotides (ATP, pink; ADP, dark red), (c) Same as panel b, viewed from the lower side and with subunit F and the VSL domains removed for clarity. (d) Type 1 binding pockets for alternating odd-numbered substrate amino acids, formed along the central channel of the Vps4 hexamer. Bound substrate amino acid side chains are shown explicitly, as are the Vps4 residues that compose the pocket (fully labeled in the second pocket). Note that the four intact amino acid binding sites form a helix around the bound ESCRT-III substrate in the central channel. Vps4 structure and color coding are the same as in panels b and c. (e) Type 2 binding pockets for alternating even-numbered substrate amino acids, formed along the central channel of the Vps4 hexamer. Bound substrate amino acid side chains are shown explicitly, as are the Vps4 residues that compose these pockets. Note that these binding sites form a second helix around the bound ESCRT-III substrate in the central channel. Vps4 structure and color coding are the same as in panels b and c. (f) Proposed mechanism of ESCRT-III translocation by Vps4. This panel shows the central translocation pore with representative Vps4 residues from the type 1 pocket (W206) and the type 2 pocket (M207) and with the ESCRT-III substrate (green) passing through the pore. Substrate translocation is proposed to occur as Vps4 subunits progress through states A to E while maintaining contacts with their respective substrate dipeptides. ATP hydrolysis at subunit D destabilizes the D/E interface and promotes displacement of subunit E toward the transitioning subunit F configuration, which allows for displacement of ADP and full release of the F subunit from the substrate (lower red arrow). Subsequent ATP binding allows subunit F to rejoin to the top of the helix (upper red arrow), where it packs against subunit A, binds the next substrate dipeptide, and assumes the subunit A configuration. Gray arrows show the relative direction of substrate peptide translocation. Panels b–f adapted from Han et al. (2017).