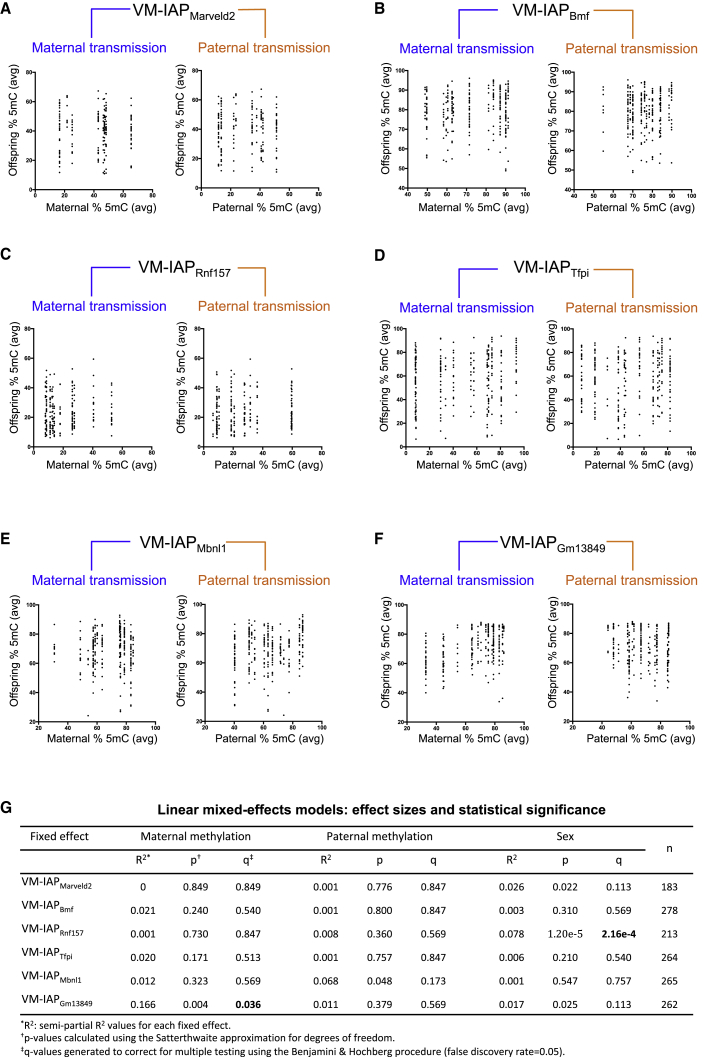
Figure 6.
Inheritance Analysis of VM-IAPs
(A–F) Methylation levels at VM-IAPs were quantified from ear samples taken from C57BL/6J breeding pairs and their offspring. Offspring methylation level is plotted against maternal and paternal methylation level for VM-IAPMarveld2 (A), VM-IAPBmf (B), VM-IAPRnf157 (C), VM-IAPTfpi (D), VM-IAPMbnl1 (E), and VM-IAPGm13849 (F). Methylation levels represent averages across the first four distal CpG sites of the VM-IAP 5′ LTRs.
(G) Statistical output from LMMs showing a significant maternal effect on offspring methylation at VM-IAPGm13849 as well as a significant sex effect on VM-IAPRnf157 methylation levels. Analyses were carried out using the lmer() function from the lme4 R package with maternal methylation level, paternal methylation level, and sex treated as fixed effects. Breeding pair and litter were treated as random effects. Raw p values were generated using the Satterthwaite approximation for degrees of freedom. Adjusted p values (q) were generated using the Benjamini and Hochberg correction to account for multiple testing. Semi-partial R2 values, representing the effect size for each fixed effect, were calculated using the r2beta() function from the r2glmm R package. Sample sizes are shown.
See also Figure S5.