Figure 2. The N-terminal Domain of an RT-Cas1 Fusion Protein is a Functional Cas6 Nuclease.
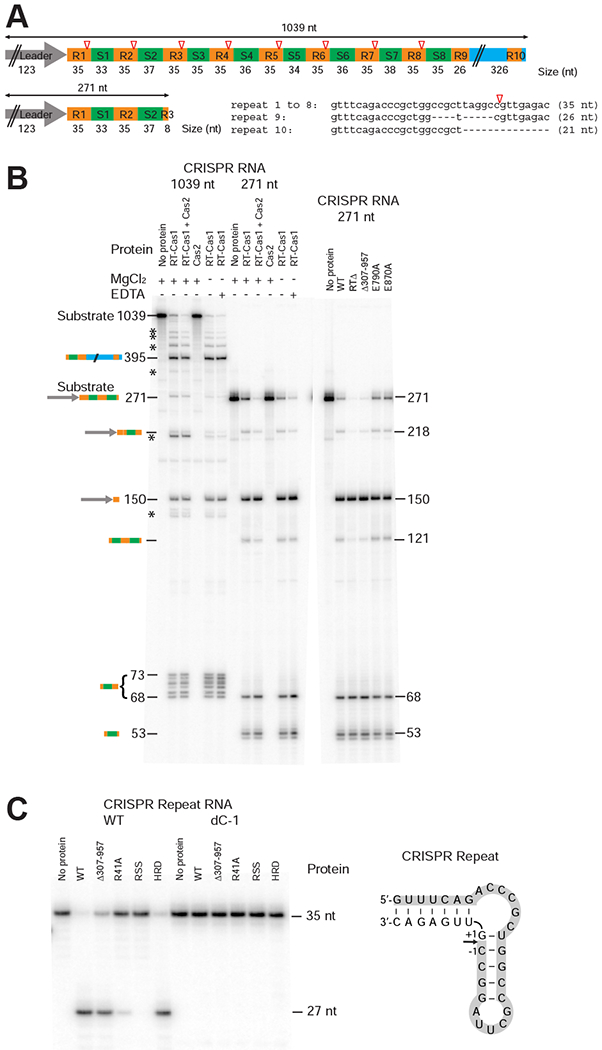
(A) Schematic of the MMB-1 CRISPR03 array transcripts used as substrates for assaying Cas6 activity. The CRISPR array contains a 123-bp leader (gray arrow), 8 identical 35-bp repeats (R1-8; orange), and 8 spacers (S1-8; 33 to 38 bp, green), followed by a partial repeat (R9), and a 326-bp downstream sequence (blue) that includes a truncated repeat (R10). Open red triangles indicate Cas6 cleavage sites 8 nt from the 3’ end of each repeat. (B) Cas6 assays. Reactions were performed using purified WT and mutant proteins (200 nM) and 32P-labeled pre-crRNA in vitro transcripts encompassing the entire 1,039 nt CRISPR03 array (2.75 nM; left) or the first 271-nt of the array from the leader through the first 8 bp of R3 (10 nM; right). The products were analyzed on a denaturing 6% polyacrylamide gel; the identities and sizes (nt) of processed pre-crRNA products are indicated to the left and right of the gels. * indicates partially processed pre-crRNAs. (C) Cas6 activity of WT and mutant Cas6-RT-Cas1 proteins assayed using CRISPR repeat-containing RNA oligonucleotide substrates. 5’-labeled RNA oligonucleotides (35-nt; 100 nM) were incubated with the indicated proteins (250 nM), and the products were analyzed on a denaturing 10% polyacrylamide gel. The sequence and predicted secondary structure of the 35-nt WT RNA oligonucleotide is shown to the right, with shading indicating the labeled 27-nt fragment resulting from Cas6 cleavage. The dC-1 RNA oligonucleotide has a deoxy C residue at position −1 from the cleavage site. The R41A, RSS (R41A, S46A, S51A), and HRD (H196A, R197A, D200A) mutants are described later in the text.
