Figure 5. Pre-crRNA Processing and CRISPR Adaptation by WT and Mutant Cas6-RT-Cas1 Proteins In Vivo.
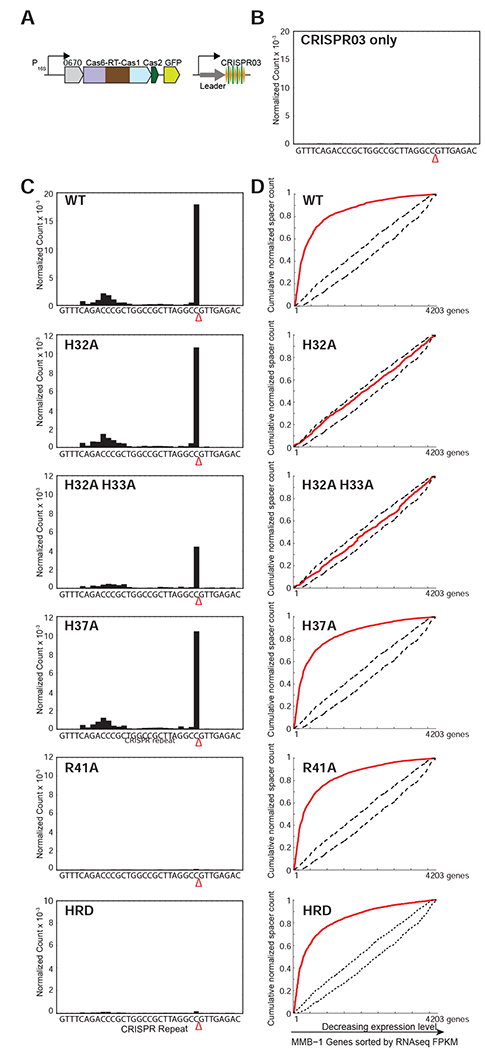
(A) Schematic of type III-B adaptation components (Cas6-RT-Cas1, Cas2, Marme_0670, along with GFP, and the CRISPR03 array) supplied on plasmid pKT230. Experiments were performed in the M. mediterranea MMB-1 III-B OperonΔ strain from which the endogenous type III-B CRISPR-Cas system was deleted, and assays were replicated with two independent transconjugant strains for each mutant. (B) Pre-crRNA processing in an empty vector control in which CRISPR array is supplied on a plasmid without type III Cas proteins. No processed crRNAs were detected by high-throughput small RNA sequencing. (C) Pre-crRNA processing by WT and mutant Cas6-RT-Cas1 proteins. WT and mutant Cas6-RT-Cas1 proteins and the CRISPR03 array were supplied on plasmid pKT230 (see above). The graphs show processed crRNA levels assayed by high-throughput small RNA sequencing. Counts are normalized to isoleucine tRNA levels (consistently the most abundant species encountered) and each experiment includes data from two independent transconjugants. The presence of a distinct 3’ end sequence in the population of CRISPR-repeat containing RNAs indicates site-specific cleavage and processing of pre-crRNA. (D) RNA or DNA spacer acquisition by WT and mutant Cas6-RT-Cas1 proteins in vivo. Assays were performed with the same cultures used in (C). Newly integrated CRISPR spacers were mapped to the MMB-1 genome. Red lines in graphs show cumulative distributions of newly acquired spacer pools (N = 500-10,000 spacers in each pool), plotted against the MMB-1 genes they were derived from sorted by expression level. WT, H37A, R41A, and HRD variants of Cas6-RT-Cas1 show preferred acquisition from highly expressed genes, suggesting spacer acquisition from RNA. Dotted black lines show expectation from random assortment (Monte-Carlo bounds: no transcription-related bias).
