Figure 6. Interaction Between the Cas6 and RT Domains of the Cas6-RT-Cas1 Fusion Protein.
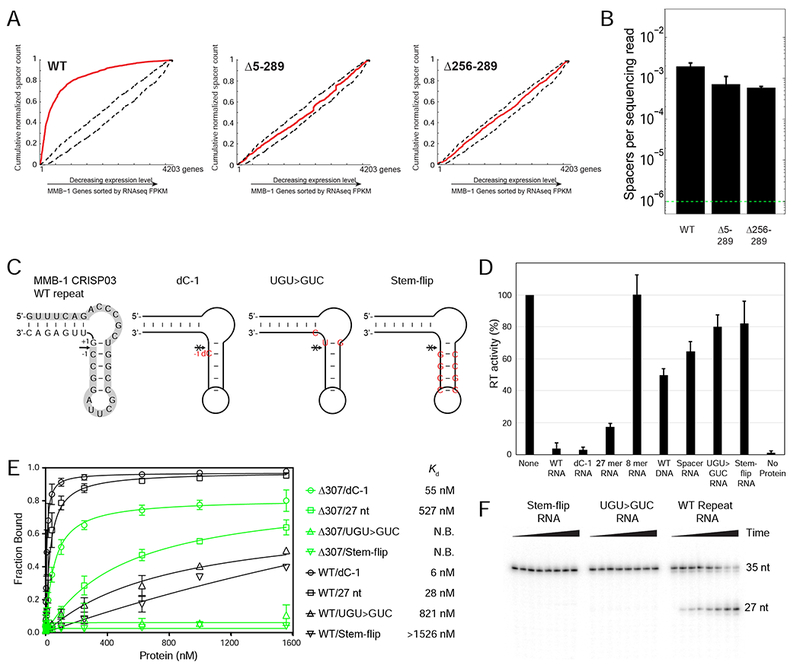
(A) In vivo CRISPR adaptation assays with WT and mutant Cas6-RT-Cas1 proteins from which the Cas6 domain (Δ5-289) or the Cas6 G-loop region (Δ256-289) were deleted. Experiments were performed as in Figure 5D, but in WT MMB-1 instead of the III-B OperonΔ genetic background. (B) Spacer detection frequency upon expression of plasmid-supplied CRISPR adaptation components as in (A). Range bars depict spacer detection frequencies for two biological replicates. The dotted green line indicates the detection limit of the assay. (C) Sequence and predicted structure of the WT and mutant MMB-1 CRISPR repeat RNA oligonucleotides with the arrow indicating the cleavage site and the 5’ 27-nt cleavage fragment highlighted in gray. The WT repeat is on the left, followed by the non-cleavable dC-1 variant, which has a single deoxy C (dC) substitution immediately upstream of the cleavage site (position −1). The UGU>GUC mutant has 3 nucleotide changes (red) that flip the G-U base-pair at the cleavage site and introduce a U to C mutation at position +2 from the cleavage site. The stem-flip variant of the CRISPR repeat has 4-bp in the stem changed to their complements (red). (D) RT activity measured by polymerization of 32P-dTTP using poly(rA)/oligo(dT)24 substrate in the presence or absence of 45 μM of WT or mutant CRISPR repeat RNA oligonucleotides. Activities are expressed as percent of a parallel no-oligonucleotide control (“None”). The bar graphs show the mean for three independent experiments with the error bars indicating the standard deviation. (E) Nitrocellulose filter binding assays for binding of WT Cas6-RT-Cas1 (black) or the isolated Cas6 domain (Δ307-957; green) to WT or mutant CRISPR repeat RNA oligonucleotides. The data were fit to a one-site binding model, and the Kd was obtained from the fit. N.B. denotes no binding. The experiment was repeated three times, with the range bars indicating the standard deviation. (F) Cas6 cleavage assays with WT and mutant CRISPR-repeat RNAs. 5’-labeled RNA oligonucleotides (35-nt; 1 μΜ) were incubated with 40 nM WT Cas6-RT-Cas1 protein. Samples were taken at intervals up to 1 hr, and products were analyzed on a denaturing 12% polyacrylamide gel to detect the appearance of the 5’-labeled 27-nt fragment.
