Figure 8: Drug-target and drug-drug interactions.
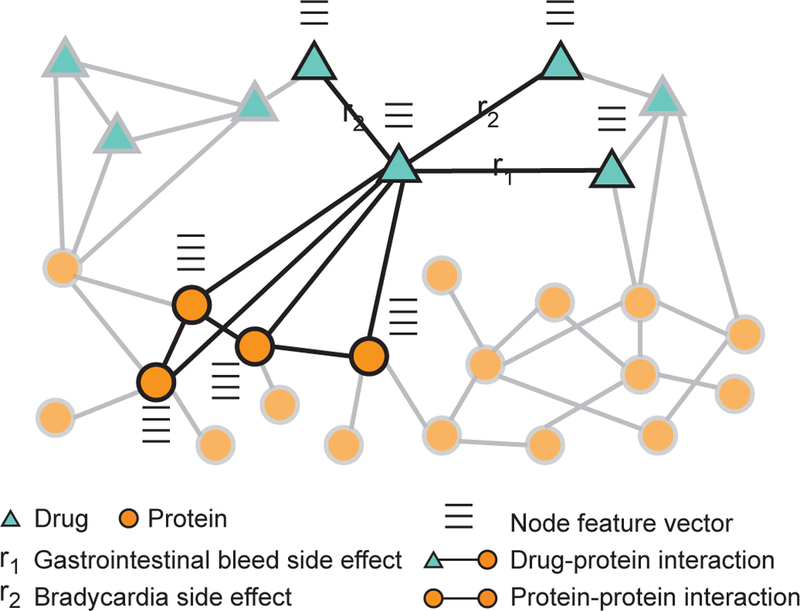
A heterogeneous network representation of drugs and proteins targeted by the drugs. In addition to interaction information, e.g., drug-drug interactions, drug-protein interactions, and protein-protein interactions (Section 8), each node in the network has a feature vector describing important biological characteristics of the node, e.g., drug’s chemical structure, and protein’s activity in tissues. Such networks are used to address two important tasks in computational pharmacology. The first is the prediction of drug-target interactions [260, 264, 19, 265], which are fundamental to the way that drugs work and often provide an important foundation for other tasks in the computational pharmacology. The second is the prediction of drug-drug interactions [273, 274, 270, 275], which are fundamental to modeling drug combinations and identifying drug pairs whose combination gives an exaggerated response beyond the response expected under no interaction. Zit-nik et al. [45] use heterogeneous networks, such as the one shown in the figure, and develop a graph convolutional deep network approach to predict which side effects a patient might develop when taking multiple drugs at the same time.
