Figure S5.
RNA-Seq Defines a STAT-1 and STAT-3 Molecular Phenotype in High-Fat-Fed Alb-Cre;Ptpn2fl/fl Mice, Related to Figure 4
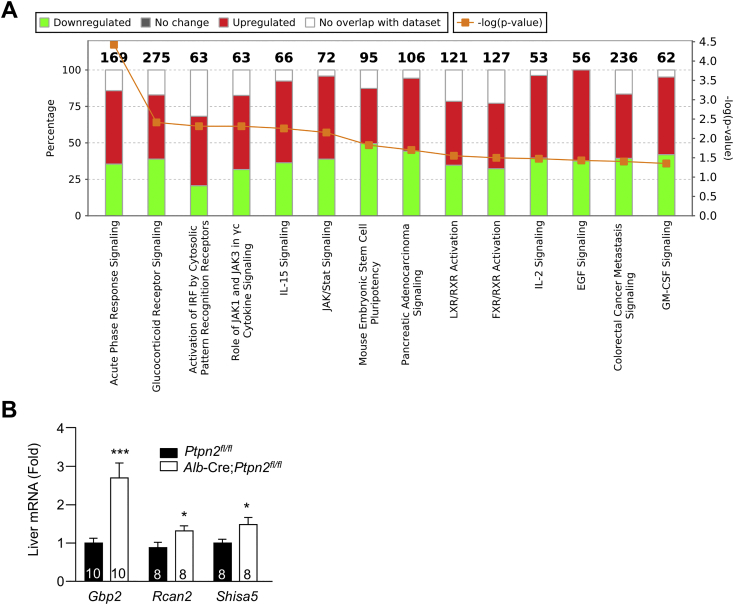
(A) Ten-twelve week-old male Alb-Cre;Ptpn2fl/fl and Ptpn2fl/fl littermate controls were fed a HFD for 40 weeks. Liver tissues from six Ptpn2fl/fl and six Alb-Cre;Ptpn2fl/fl mice were processed for transcriptome analysis by RNaseq. Stacked bar-chart depicting the overlap between the top 119 differentially expressed hepatic genes in Alb-Cre;Ptpn2fl/fl mice by RNA-seq (log fold-change > 0.2 and p value < 0.01) and annotated Canonical Pathways in the Ingenuity Knowledge Base. Numbers (top) indicate the total number of genes included in the pathway. Red, green and clear bars indicate overlap with upregulated genes, overlap with downregulated genes, and genes not in the regulated set, respectively (left-hand axis). p values for the extent of overlap of gene sets (orange line; scaled to negative log10, see right-hand axis) were calculated using a right-tailed Fisher’s exact test.
(B) An independent cohort of ten week-old male Ptpn2fl/fl and Alb-Cre;Ptpn2fl/fl mice were fed a HFD for 40 weeks and liver tissues processed for quantitative real time PCR. Quantified results (means ± SEM) are shown for the indicated number of mice with significance determined using a Student’s t test.