Figure 3.
Fluorescence Microscopy Gives Single-Cell Information on Locus Conformation
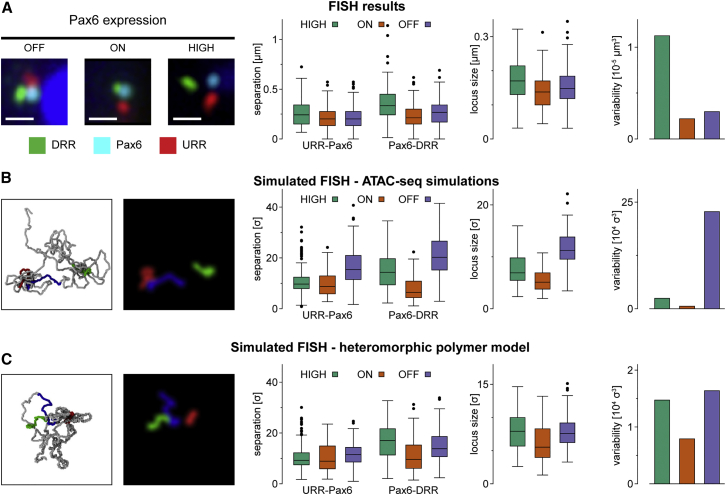
(A) 3D FISH experiment using probes positioned at the URR, DRR, and Pax6 gene (Figure S3A). Left: representative three-color 3D FISH images. Scale bar, 0.5 μm. Mid-left: distributions of probe separations shown as boxplots. Mid-right: boxplots showing the size of the locus calculated from three-color FISH experiments. Right: bar graph displaying variability in locus conformation (see STAR Methods and Figure S3E). In general, the locus becomes more compact in Pax6-ON cells compared to Pax6-OFF cells but becomes less compact (and more variable) in Pax6-HIGH cells.
(B) Simulated FISH data extracted from locus conformations generated by chromatin folding simulations using ATAC data to define TF-binding sites (as shown in Figures 2D and 2E). Left: representative snapshot of the locus conformation alongside a simulated FISH image shown for illustrative purposes. Mid-left: boxplots showing distances between probes given in simulation units (σ) (see STAR Methods). Right: locus size and structural variability as in (A), but data are given in simulation units. These simulations depart from the experimental measurements, as the Pax6-OFF cells are the most decompacted and highly variable.
(C) Simulated FISH measurements from the heteromorphic polymer model. Agreement with the experimental data improves in that the locus is decompacted in Pax6-HIGH cells and the variability is far larger in the highly expressing cells.
See also Figure S3.