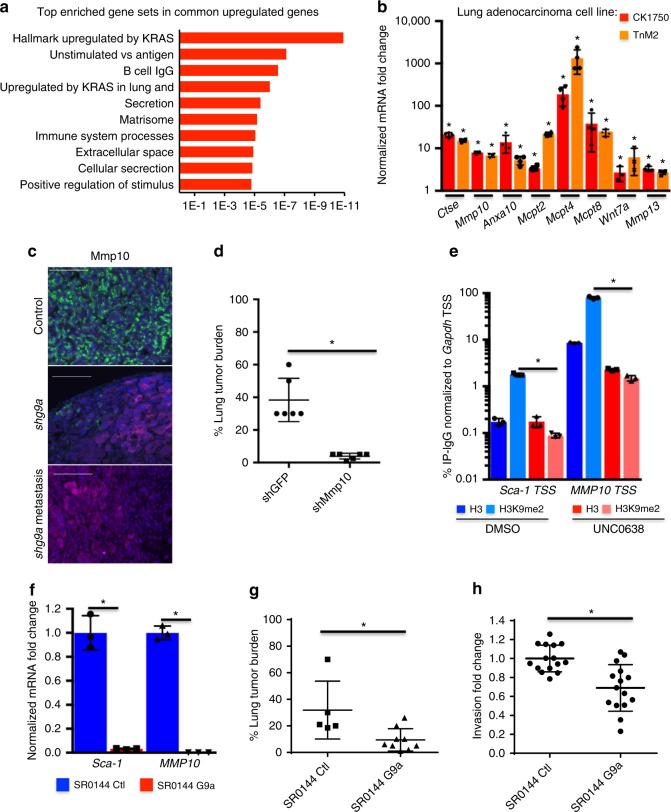
Fig. 3.
G9a controls TPCs through KRAS-associated and ECM-associated genes. a Molecular Signatures Database enrichment of gene terms associated with genes upregulated upon G9a/Glp inhibition. b Bar chart of fold change of Gapdh-normalized mRNA levels of indicated genes in adenocarcinoma cells treated with 1 µM UNC0638 relative to DMSO-treated controls. Error bars denote standard deviation. *P < 0.05, Fisher’s t test, n = 4. c Representative (n = 5) images of shLuc control, and shG9a lung tumors and shG9a metastases immunostained for the indicated proteins. Scale bar = 100 µm. d Chart of % lung tumor area per H&E-stained section from recipients of intravenously injected shGFP, and shMmp10 SR0144 shG9a lung adenocarcinoma cells. *P < 0.05, Fisher’s t test. e Representative bar chart of % immunoprecipitation compared to input DNA–IgG of H3 and H3K9me2 ChIPs at the indicated TSS relative to Gapdh TSS of lung adenocarcinoma cells treated with 1 µM UNC0638 or DMSO. Error bars denote standard deviation. *P < 0.05, Fisher’s t test, n = 3 technical replicates. f Bar chart of mRNA from the indicated genes normalized to Gapdh from control and G9a-overexpressing SR0144 shG9a lung adenocarcinoma cells. Error bars denote standard deviation, *P < 0.05, Fisher’s t test, n = 3. g Chart of % lung tumor area per H&E-stained section from recipients of intravenously transplanted control and G9a-overexpressing SR0144 shG9a lung adenocarcinoma cells. *P < 0.05, Fisher’s t test. h Chart of fold change in invasion through a 3D matrix-coated migration transwell of G9a-overexpressing relative to control SR0144 shG9a lung adenocarcinoma cells. *P < 0.05, Fisher’s t test