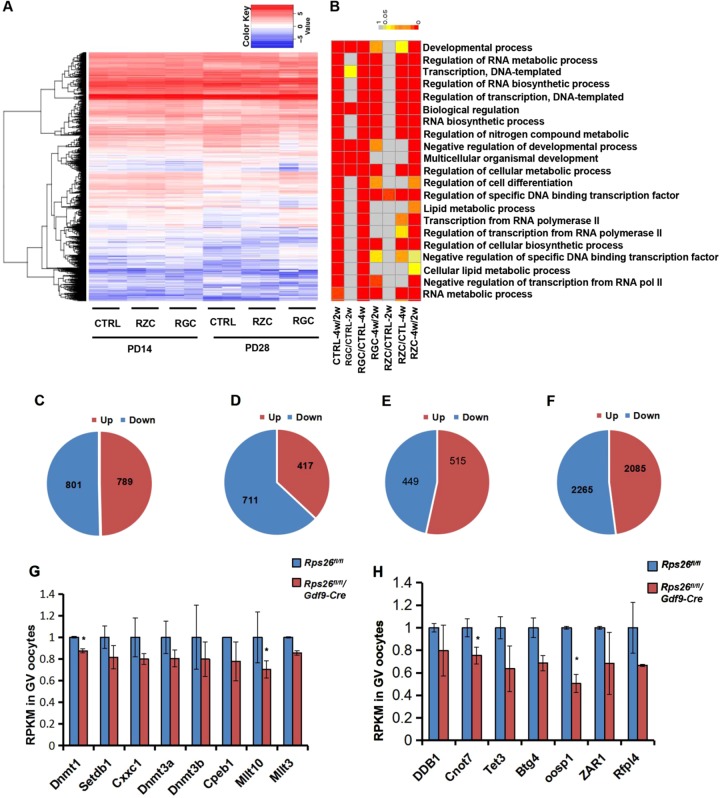
Fig. 5. Rps26 regulates the mRNA transcription involved in methylation modifications and developmental processes.
a Heatmap of mRNA expression in oocytes showing changes in a subset of genes after Rps26 deletion in Rps26fl/fl/Zp3-Cre (RZC) and Rps26 fl/fl/Gdf9-Cre (RGC) mice compared with the control (CTRL) Rps26fl/fl mice at PD14 and PD28. b Gene ontology showing the biological processes affected by the altered mRNA expression by comparing the gene expression in the different groups of oocytes at PD14 (2w) and PD28 (4w).c–d Numbers of upregulated (up) genes and downregulated (down) genes in oocytes of control (CTRL) Rps26fl/fl mice (c) and Rps26 fl/fl/Gdf9-Cre mice (d) at PD28 compared to PD14. e–f Numbers of upregulated (up) genes and downregulated (down) genes in oocytes of Rps26 fl/fl/Gdf9-Cre mice compared to Rps26fl/fl mice at PD14 (e) and PD28 (f). g Genes of histone methyltransferases of H3K4 or H3K9 and other related genes were downregulated in Rps26 fl/fl/Gdf9-Cre mice compared to Rps26fl/fl mice as determined from RNA sequencing. *p < 0.05 as calculated by two-tailed Student’s t-tests. Genes involved in DNA methylation and oocyte-specific genes were mostly downregulated in Rps26 fl/fl/Gdf9-Cre mice compared to Rps26fl/fl mice as determined by RNA sequencing. *p < 0.05 as calculated by two-tailed Student’s t-tests