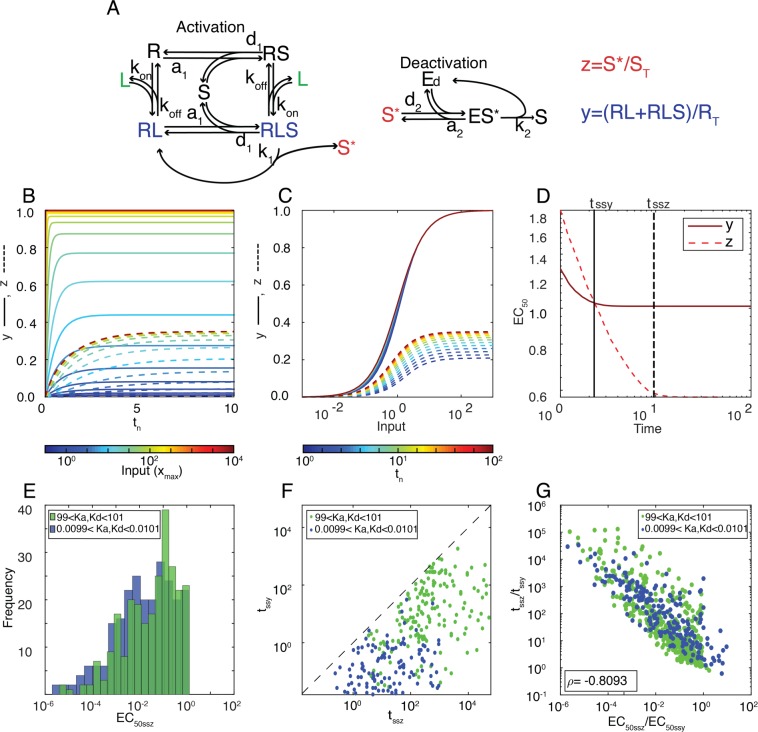
Figure 4.
The LR-CM system is a better shifter than either in isolation. (A) Reaction scheme of a biological network composed of a LR coupled to a CM cycle. The receptor has four possible states: unbound (R), bound to the ligand (RL), bound to the substrate (RS), and bound to both (RLS). RLS is also the enzyme-substrate complex that catalyzes S activation into S*. The enzyme Ed forms a complex with S*, ES* to deactivate it into S, completing the CM cycle. (B,C) For an example parameter set, plot shows the time course and input-output curves (y = (RL = RLS)/RT), solid lines and z = (S*/ST), dashed lines) at different times distributed uniformly in log scale from t0 = 1 to tf = 102 (using a heatmap color scale). Each curve is normalized by its maximum value at steady-state. (D) For the same example as in (B and C) EC50 vs time for both outputs. Vertical lines mark the time tss at which each curve reached ss. Parameter set used in (B–D) a1 = a2 = d1 = d2 = k1 = k2 = ET = ST = RT = 1. Binding rates a1 and a2 are in units of ([time] [concentration])−1, unbinding rates d1, d2, k1 and k2 are in units of [time]−1 and total amounts ET, ST and RT in units of [concentration]. (E) EC50ssz for 200 parameter sets sampled randomly within the ranges defined in Table S1 so that Ka [((d1 + k1)/(a1 * ST))] and Kd [((d2 + k2)/(a2 * ST))] fell in the ranges indicated. (F) tssy vs. tssz for the 200 sets shown in (E). (G) The relative speed of the two modules tssz/tssy negatively correlates with the relative sensitivity (EC50ssz/EC50ssy).