Figure 1.
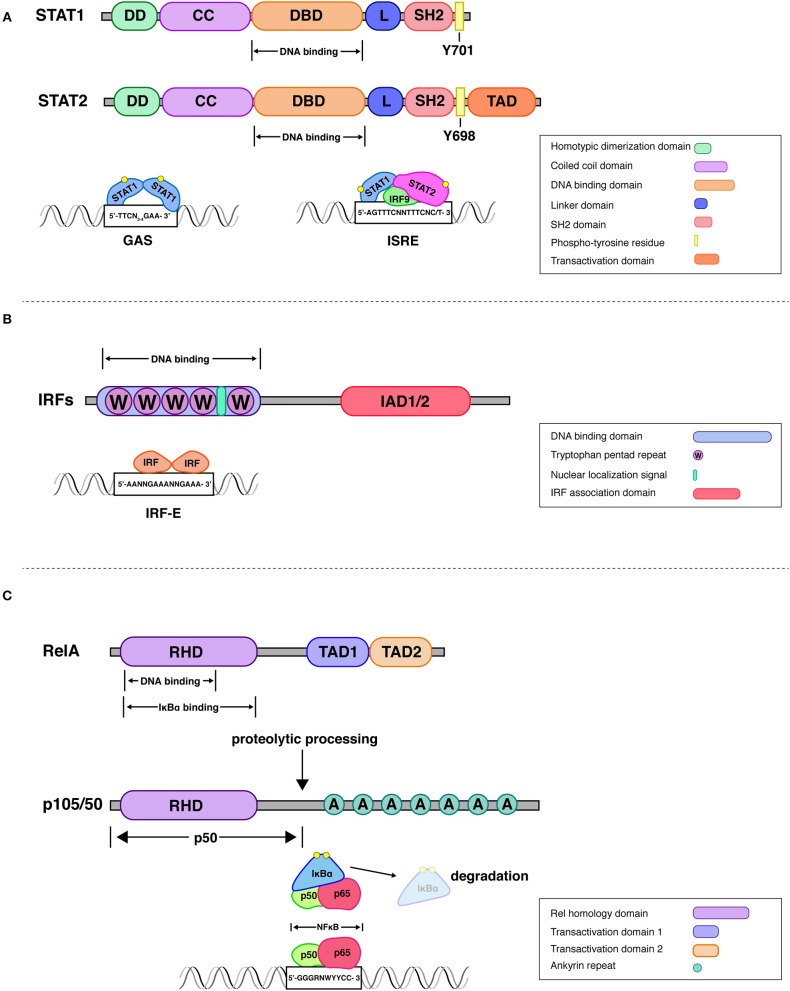
Structural attributes of STAT-, IRF-, and Rel family transcription factors. STATs. All mammalian STAT members share a common structural motif consisting of an N-terminal domain, which plays a role in dimerization (DD), followed by a coiled-coil domain (CC), that can be involved in interactions with other proteins, a DNA-binding domain (DBD), a linker domain (L), an SH2 domain for reciprocal phospho-tyrosine interaction and a transactivation domain (10, 11). Upon receptor engagement Janus kinases lead to the activation of the latent cytoplasmic STATs, via phosphorylation on single tyrosine residues (Y701 on STAT1 and Y690 on STAT2). The STAT1-STAT2 dimer associates with interferon regulatory factor 9 (IRF9) to form a transcriptionally active IFN-stimulated gene factor 3 (ISGF3). This complex controls gene expression by binding to interferon-stimulated response elements (ISRE) present in promoters of IFN stimulated gene (ISG). Additionally, STAT1 homodimers, translocate to the nucleus and stimulate ISG expression by binding to gamma interferon-activated sites (GAS) (17). IRFs. All IRFs harbor a conserved N-terminal DNA-binding domain (DBD), which forms a helix-turn-helix domain with a conserved tryptophan cluster that recognizes DNA sequences in interferon induced genes (18). An analysis of the crystal structure of the DBD of IRF1 bound to the Ifnb promoter revealed that 5′-GAAA-3′ is the consensus sequence recognized by the helix-turn-helix motif of IRF1 (19). This DNA motif is known as the IRF-element (IRF-E) (20). All IRFs harbor a C-terminal IRF association domain (IAD), which is responsible for homo- and heteromeric interactions with other family members or transcription factors (21, 22). IAD1 and IAD2 domains can be distinguished by structural criteria and are found, respectively, in IRF1 and IRF2 or all other IRFs. Rel (NFκB). One of the best studied NFκB dimers is the p50/p65 heterodimer, whose crystal structure has been solved (23). NFκB recognizes 9–11 bp (base pair) DNA-elements, which are often located within promoters and enhancers of NFκB target genes. The consensus sequence 5′-GGGRNWYYCC-3′, where R denotes a purine base, N means any base, W stands for adenine or thymine and Y represents a pyrimidine base, is recognized by the Rel-homology domain [RHD; (12)]. The C-terminal domain of RelA (p65) contains two strong and independent transactivation domains (TAD) providing full transcriptional activity (24). The p100 precursor protein is proteolytically processed to the NFκB subunit p50. The mature p50 protein contains the RHD followed by glycine-rich region, a region that is essential for directing the cleavage and proteolytic processing of a long IκB-like C-terminal part of the precursors (25). IκBα regulates rapid and transient induction of NFκB activity. The crystal structure of IκBα bound to the p65/p50 heterodimer revealed that one IκBα molecule binds to an NFκB dimer and masks the NLS of p65. IKKβ is necessary and sufficient for phosphorylation of IκBα, leading to IκBα ubiquitination, and further degradation by the proteasome.