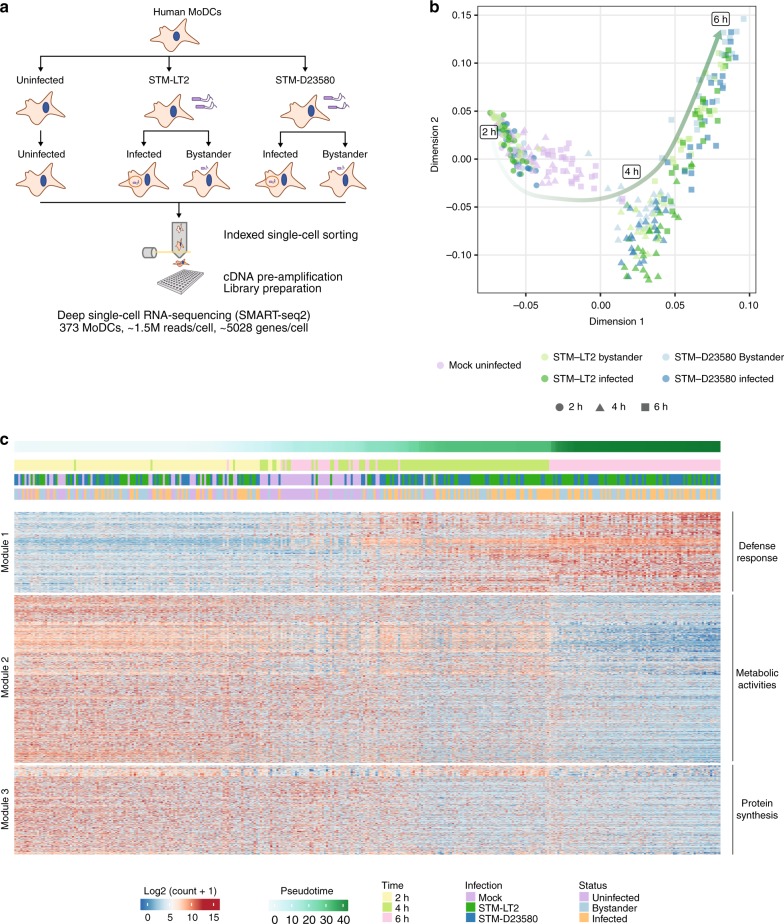
Fig. 1.
Single-cell transcriptomics analysis of human MoDCs challenged with invasive or non-invasive Salmonella. a Schematic representation of the experimental design. Human MoDCs were challenged with labelled STM-LT2 or STM-D23580 bacterial strains. At 2, 4 and 6 h post infection (p.i.), infected and bystander cells were sorted and processed according to the Smart-seq2 protocol. b The diffusion map non-linear dimensionality reduction technique was applied on the first 50 components of a principal component analysis (PCA) computed on the 500 most variable genes to visualize the high-dimensional relations between cells in the data set. The diffusion map reveals distinct cell populations that progress through the three time points of the experimental design. c Heat map displaying log2-transformed normalized counts for 3188 genes that significantly change with pseudotime across all single-cells (q-value < 0.001). Pseudotime was computed from a set of 2759 genes differentially expressed between Salmonella-challenged cells at 2 and 6 h p.i. (q-value < 0.001). Experimental metadata (i.e., time point, infection, status) are indicated for each cell as colour bars above the heat map