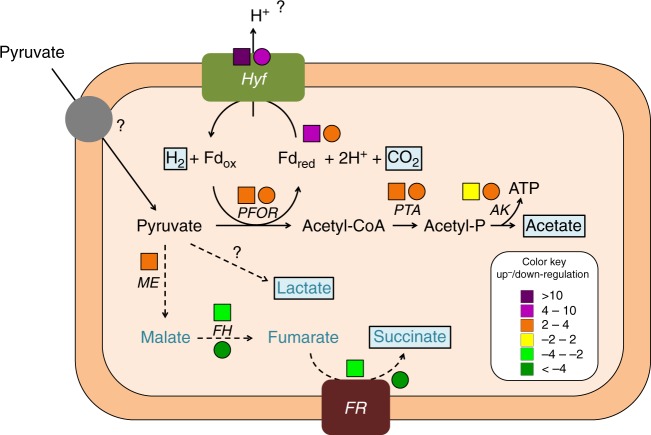
Fig. 7.
Tentative scheme of pyruvate fermentation metabolism in S. multivorans and S. cavolei. Reactions represented by solid arrows belong to the core pyruvate metabolism and are catalyzed by both organisms. Reactions with dashed arrows are solely catalyzed by S. multivorans, fumarate hydratase and fumarate reductase are also present in S. cavolei. Hyf might pump protons via its membrane-integral subunits (Supplementary Note 1, Supplementary Table 6 and Supplementary Figures 10–13) which could lead to additional ATP production via chemiosmotic coupling. Question marks indicate enzymes not identified. Fermentation products are highlighted in light blue boxes. Protein abundance ratios (pyruvate alone versus pyruvate/fumarate) are indicated by colored squares (S. multivorans) and circles (S. cavolei) at the protein abbreviations. Color code of the ratios is given in the box at the lower right. Hyf Hyf-like hydrogenase (SMUL_2383–2392; SCA02S_RS01920-RS01965), PFOR pyruvate:ferredoxin oxidoreductase (SMUL_2630; SCA02S_RS04525), PTA phosphotransacetylase (SMUL_1483; SCA02S_RS00245), AK acetate kinase (SMUL_1484; SCA02S_RS00240), ME malic enzyme (SMUL_3158; corresponding enzyme in S. cavolei is not present), FH fumarate hydratase (SMUL_1459, SMUL_1679–1680; SCA02S_RS00615-RS00620), FR fumarate reductase (SMUL_0550–0552; SCA02S_RS07735-RS07740)