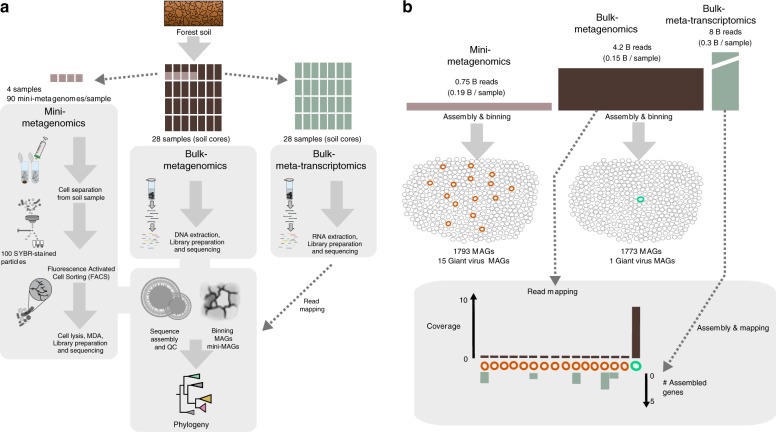
Fig. 1.
Discovery pipeline for soil giant viruses. a Overall workflow. Fourteen forest soil cores from Barre Woods long-term experimental warming site were sub-sampled into organic horizon and mineral zone resulting in 28 total samples. Total DNA and RNA were extracted from 28 soil samples for bulk metagenomics and metatranscriptomics. Of these samples, a subset of four encompassing two organic and two mineral layers were selected for flow-sorted mini-metagenomics. Cells and presumably viral particles, were separated from soil, stained with SYBR green nucleic acid stain and sorted using fluorescence activated cell sorting (FACS). Ninety sorted pools of 100 SYBR+ particles underwent lysis, whole genome amplification, library preparation, and sequencing on the Illumina NextSeq platform. Phylogenomic analysis of metagenome assembled genomes (MAGs) facilitated the identification of novel giant viruses. There was no correlation of presence or absence of giant viruses and sample treatment (Supplementary Table 3). b Data analysis summary. Fifteen giant virus MAGs (orange circles) were recovered from sorted samples, while only one giant virus MAG (turquoise circle) was recovered from the bulk metagenomes. The other 1778 MAGs from the mini-metagenomes (gray circles) and 1772 MAGs from the bulk metagenomes (gray circles) were of bacterial or archaeal origin and not analyzed further in this study. Mapping of bulk metagenome reads to MAGs revealed ~9× coverage of the bulk-metagenome derived MAG and <1× coverage of MAGs derived from mini-metagenomes, confirming the inability to recover these novel giant virus genomes using bulk metagenomics despite deep sequencing efforts. Assembly and mapping of metatranscriptome data indicated expression of only few of the novel giant virus genes of MAGs derived from mini-metagenomes