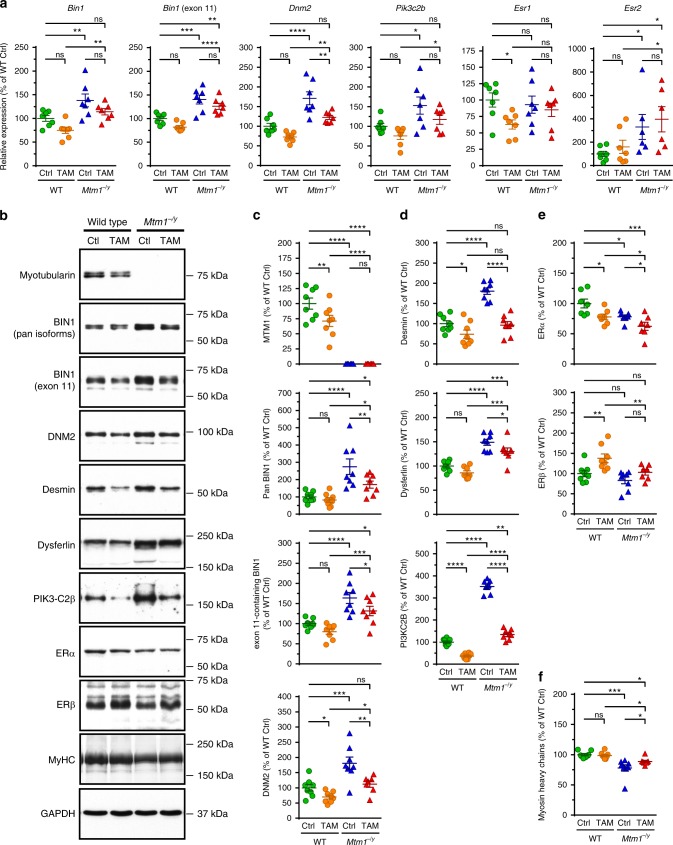
Fig. 4.
Tamoxifen reduces the expression of XLMTM disease modifiers and alters the expression of estrogen receptors. The mRNA and protein levels of known XLMTM disease modifiers and of estrogen receptors, which mediate most tamoxifen actions, were determined in gastrocnemius muscle of 42-days old (D42) wild type (WT) and Mtm1−/y mice, either untreated (control; Ctrl) or tamoxifen (TAM)-treated (30 mg kg−1 of diet). a mRNA levels normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh). From left to right, relative expression (as percentage of WT Ctrl) of mRNA encoding amphiphysin 2/BIN1 (Bin1; all transcripts), muscle-specific BIN1 (Bin1; exon 11-containing transcript), dynamin-2 (Dnm2), PI3KC2B (Pik3c2b), estrogen receptor (ER) α (Esr1), and ERβ (Esr2). b Representative immunoblots of proteins of interest, as indicated. DNM2: dynamin 2; MyHC: myosin heavy chains; GAPDH: glyceraldehyde-3-phosphate dehydrogenase. Position of molecular weight markers (kDa) is shown. c–f Levels (normalized to GAPDH and expressed as percentage of WT Ctrl) of proteins selected for their role in XLMTM and tamoxifen signaling. c Proteins involved in the “MAD”-pathway. From top to bottom: myotubularin (MTM1), amphiphysin 2/BIN1 (pan-isoforms), amphiphysin 2/BIN1 (muscle-specific isoform), dynamin-2 (DNM2). d Other disease modifiers and protein deregulated in absence of MTM1. From top to bottom: desmin, dysferlin, PI3KC2B. e Estrogen receptors. Top: ERα; bottom: ERβ. f Myosin heavy chains, a major constituent of sarcomeres. Data represent the mean ± s.e.m. of n = 6–8 muscles per group. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns non-significant. One-way ANOVA followed by Fisher’s LSD post-test