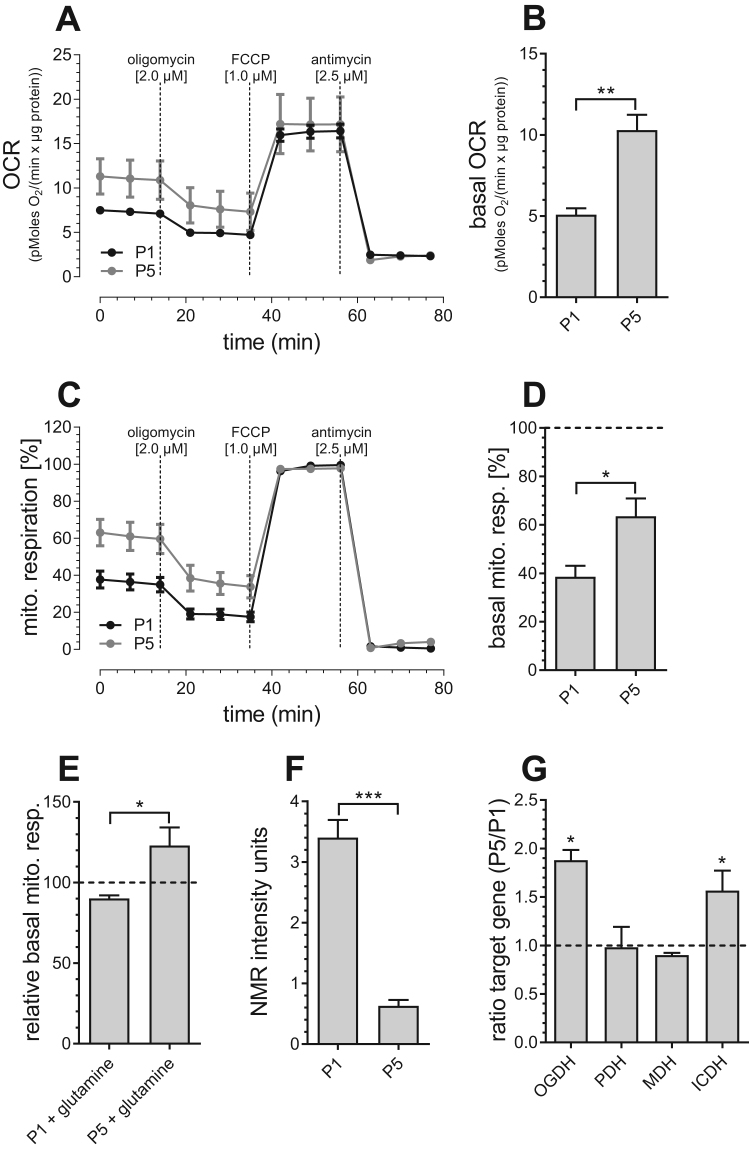
Fig. 2.
Mitochondrial metabolism in P1 and P5 PAECs. Mitochondrial respiration of P1 (black curve) and P5 (gray curve) PAECs in response to oligomycin [2 µM], FCCP [1 µM] and antimycin [2.5 µM] normalized to protein content (A). Bar graphs show basal mitochondrial respiration before addition of compounds (B). Mitochondrial respiration of P1 (black curve) and P5 (gray curve) PAECs in response to oligomycin [2 µM], FCCP [1 µM] and antimycin [2.5 µM], normalized to maximal mitochondrial respiration (C). Bar graphs show basal mitochondrial respiration presented as percentage of respective maximum mitochondrial respiration (D). Basal mitochondrial respiration of P1 and P5 PAECs after 2 mM glutamine treatment, presented as percentage of basal mitochondrial respiration of respective untreated control cells (E). Metabolite levels depicted as normalized NMR intensity units (a.u.) (F). mRNA expression ratios of OGDH, PDH, MDH and ICDH in P5 versus P1 PAECs (G). Data are representative of ≥ 3 biological repeats ± SEM. Significant differences were assessed via unpaired t-test and presented as specific p-values (* = p ≤ 0.05, ** = p ≤ 0.01, *** = p ≤ 0.001).