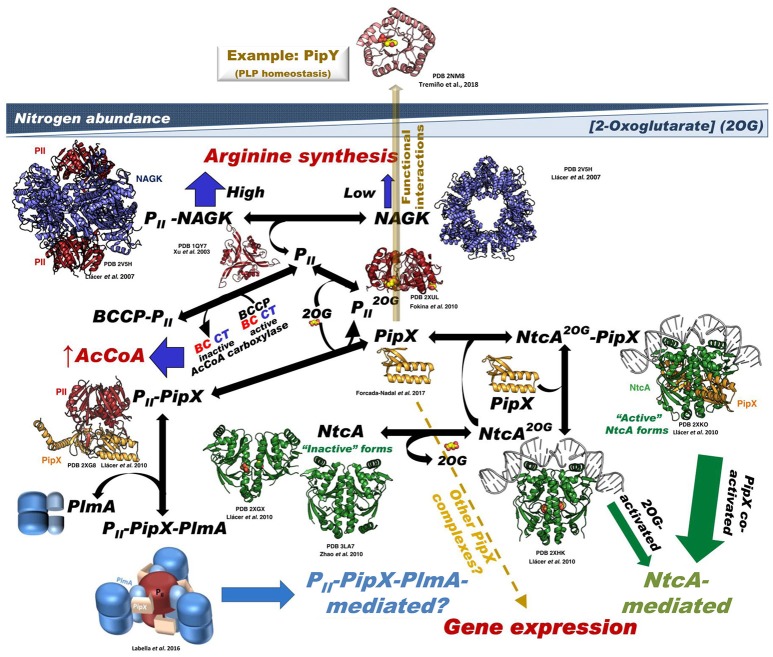
Figure 1.
Summary of the PII-PipX-NtcA network of S. elongatus. The network illustrates its different elements and complexes depending on nitrogen abundance (inversely related to 2OG level) and the structures of the macromolecules and complexes formed (when known). For PlmA (dimer in darker and lighter blue hues for its dimerization and DNA-binding domains, respectively) and its complex the architectural coarse model proposed (Labella et al., 2016) is shown, with the C-terminal helices of PipX (schematized in the extended conformation) pink-colored and the two PII molecules in dark red. The DNA complexed with NtcA and with NtcA-PipX is modeled from the structure of DNA-CRP (Llácer et al., 2010), since no DNA-NtcA structure has been reported. BCCP, biotin carboxyl carrier protein of bacterial acetyl CoA carboxylase (abbreviated AcCoA carboxylase); the other two components of this enzyme, biotin carboxylase and carboxyl transferase are abbreviated BC and CT, respectively. No structural model of BCC has been shown because the structure of this component has not been determined in S. elongatus and also because the structures of this protein from other bacteria lack a disordered 77-residue N-terminal portion that could be highly relevant for interaction with PII. The yellow broken arrow highlights the possibility of further PipX interactions not mediated by NtcA or PII-PlmA resulting in changes in gene expression (Espinosa et al., 2014). The solid semi-transparent yellowish arrow emerging perpendicularly from the flat network symbolizes the possibility of functional interactions of PipX not mediated by physical contacts between the macromolecules involved in the interaction, giving as an example the functional interaction with PipY. Its position outside the network tries to express the different type of interaction (relative to the physical contacts shown in the remainder of the network) as well as to place it outside the field of 2OG concentrations.