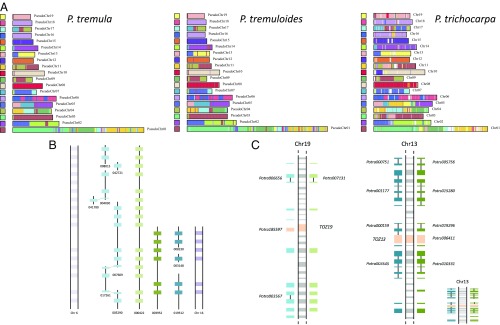
Fig. 2.
Genome synteny. (A) Self-alignments of nucleotide sequences for the P. tremula, P. tremuloides, and P. trichocarpa genomes, showing that paralogous regions from the Salicaceae WGD event are largely retained across the three genomes. Synteny matches following a WGD event are indicated by colored blocks. (B) Schematic representation of scaffold Potra00422, with genes shown as light green boxes, and orthologs in P. tremuloides (light blue) and P. trichocarpa (light purple), illustrating an example of retained local synteny between the three genomes. Paralogs are colored in darker shades of each color (to the right side in the representation). Dotted lines between some of the genes in P. tremuloides indicate that the gene is split across different scaffolds. (C) Schematic representation of the sex-determining region in aspen. (Left) Middle shows TOZ19 and 12 genes upstream and downstream. On both sides, the orthologs (detected using both BLAST and conserved synteny) in P. tremula and P. tremuloides are depicted. (Right) TOZ19 paralog, TOZ13, accompanied by 12 genes on each side. Scaffold IDs are noted for the TOZ19 and TOZ13 genes, as well as in cases of more than one gene per scaffold. Dotted lines indicate paralogs in P. trichocarpa. (Inset, Bottom Right) Region on chromosome 13 drawn to the same scale used for the region on chromosome 19.