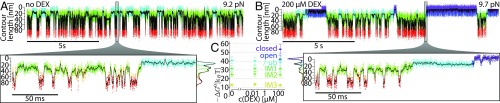
Fig. 3.
GR folds through many on-pathway intermediates before it binds hormone and reaches the native state. (A) Contour length vs. time traces in passive-mode without DEX added in solution. The trace is colored according to HMM state assignment (details in text and SI Appendix). The given force bias refers to the force acting on the closed state. A zoom into the data is shown below. Next to the zoom is a histogram of the smoothed contour length data with the contributions of the subpopulations highlighted in color. (B) Measurement similar to that in A, but with 200 µM DEX in solution. (C) Comparison of free energy differences of the states to the unfolded state at different hormone concentrations. The error bars combine statistical as well as the estimated systematic error of the force calibration.