Abstract
Shiga toxin-producing Escherichia coli (STEC) O157 and non-O157 are food-borne pathogens and contaminants of foods of animal origin. This study was conducted to investigate the presence of virulence and integrase genes in STEC isolates from diarrhoeic calves in Fars Province, Iran. Five hundred and forty diarrheic neonatal calves were randomly selected for sampling. Rectal swabs were collected and cultured for isolation and identification of E. coli following standard methods. The isolates were analysed for the presence of class 1 integrons and bacterial virulence factors using polymerase chain reaction (PCR). Antimicrobial susceptibility testing was performed using the Kirby–Bauer disc diffusion method. Out of 540 diarrhoeic faecal samples, 312 (57.7%) harboured E. coli and 71 (22.7%) of them were identified as STEC: 41(69.5%) carried the stx2 gene, 21 (35.6%) carried the stx1 gene and 3 (5%) carried both. Twenty-six (44%) of the isolates showed the eae gene. Among the STEC isolates examined for susceptibility to eight antimicrobial agents, erythromycin and penicillin (96.8%) resistance were most commonly observed, followed by resistances to ampicillin (71.8%), tetracycline (62.5%) and trimethoprim/sulfamethoxazole (39%). Integrons were detected by PCR in 36% of the STEC tested isolates, 57 (89%) of which showed resistance to at least three antimicrobial agents. Our findings should raise awareness about antibiotic resistance in diarrhoeic calves in Fars Province, Iran. Class 1 integrons facilitate the emergence and dissemination of multidrug-resistance (MDR) among STEC strains recovered from food animals.
Introduction
New research provides the strongest evidence that Shiga toxin-producing Escherichia coli (STEC) non-O157:H7 and in particular serogroup O157 are linked to severe gastrointestinal diseases (Dehkordi et al. 2014). The clinical manifestations of STEC infection can vary, from asymptomatic carriage to very serious illnesses such as haemolytic uremic syndrome (HUS), thrombocytopenic purpura (TTP) and haemorrhagic colitis (HC) (Thomas et al. 2012). Estimates vary, but experts suggest that gastrointestinal infections are responsible for approximately 1.5 million deaths per year, over 90% of which are in developing countries (Montenegro et al. 2011). For instance, non-O157:H7 serogroups are found in more than 36 000 cases of infections annually and at least 73 000 are infected with O157:H7 serogroup in the United States (US) (Zhao et al. 2001).
Studies have revealed that O157 and non-O157 strains of cattle origin can cause the disease in humans via consumption of raw milk and undercooked meat. In fact, cattle, especially young animals, are known to be the primary reservoirs of both non-O157 and O157 STEC (Moura et al. 2012). The pathogenicity of STEC is associated with Shiga toxin (stx) encoded by Shiga toxinogenic (stx) genes 1, 2 (stx1 and stx2) and an outer membrane protein which is encoded by the chromosomal eae gene (Pradel et al. 2008).
The problems with some new STEC strains isolated from neonatal calf diarrhoea (NCD) (Rigobelo et al. 2008), which is recognised as a disease complex characterised by acute, undifferentiated diarrhoea in newborn calves, are that antibiotic multiresistance (De Verdier et. al. 2012) and STEC strains can be transmitted to humans by contact occupational exposure and the food chain (Schroeder et al. 2002). Epidemiological observations show high levels of antimicrobial resistance in bacterial pathogens from veterinary and human medicine (Zhao et al. 2001). This has led to the discovery that these bacteria are able to acquire antibiotic resistance by resistance-conferring genes, many of which are carried on transposons, plasmids or integrons (Bakhshi, Najibi & Sepehri-Seresht 2014). An integron is mainly composed of an integrase gene that encodes a site-specific recombinase, by which an insertion site of integron is recognised. Moreover, an integron contains a variable region which is the place for gene cassettes to be inserted (White et al. 2001). Depending on the sequence of the encoded integrases (intI) catalysing excision and integration of deoxyribonucleic acid (DNA) units, eight distinct integron classes have been identified up to now, and class 1 integrons have shown to be the major contributors to multidrug-resistant (MDR) infections in the Enterobacteriaceae family (Singh et al. 2005).
Many studies in various countries including Iran have shown that the distribution of integrons among enteric bacteria has increased over time (Eftekhari et al. 2013; Gonzalez et al. 1998; Hamada, Oshima & Tsuji 2003; Martinez-Freijo et al. 1998, 1999; Najibi et al. 2012). In Iran, only a few studies have reported antimicrobial resistance properties and virulence genes in the pathogenic E. coli (Bakhshi et al. 2014; Shahrani et al. 2014). Unfortunately, there is no conclusive data on the distribution of virulence genes and the antimicrobial resistance properties of STEC strains isolated from Iran, particularly from Fars, which is one of the major agricultural and animal husbandry areas in Iran, with nearly 400 000 cattle and 8 000 000 sheep and goats (Shams et al. 2012).
Materials and methods
Study design and study areas
Sampling and Escherichia coli identification
A total of 540 recto-anal mucosal swabs from diarrhoeic calves (< 30 days of age) were collected over 1 year from November 2015 to November 2016.
These calves were raised on 33 farms from eight geographic areas in Fars Province, including industrial, semi-industrial and traditional farms, with a herd size of 25–500 cows. These farms had a recognised scouring problem in neonatal calves. Sick calves which showed abnormal faecal consistency and/or signs of dehydration and weakness were selected. None of them had been vaccinated. All samples were immediately placed in cooled boxes and transported to the laboratory. The swab samples were incubated overnight at 37 °C in trypticase soy broth (TSB) (Merck KgaA, Darmstadt, Germany). Each sample was then streaked onto MacConkey’s agar (MC, Merck, Germany) (24 hours at 37 °C). Lactose positive colonies were cultured on eosin methylene blue agars (EMB, Merck, Germany) (24 h at 37 °C). Green colonies with a metallic lustre were considered typical E. coli colonies. Such colonies were confirmed as E. coli using standard biochemical tests (citrate utilisation, indole production, glucose, lactose fermentation, urease negative and hydrogen sulphate production). The biochemically confirmed E. coli colonies were subjected to DNA analysis.
Antimicrobial susceptibility and multidrug resistance
Antimicrobial susceptibility testing against eight antimicrobials was performed on 52 O157 and 12 non-O157 STEC isolates using the disk diffusion method on Mueller Hinton agar plates (Merck, Germany) based on the Clinical and Laboratory Standards Institute (CLSI) guidelines (Wayne 2012a). The following antibiotics (PadtanTeb, Iran) were applied: chloramphenicol (C: 30 µg), erythromycin (E: 25 µg), ampicillin (AM: 10 µg), trimethoprim/sulfamethoxazole (SXT: 30 µg), penicillin (P: 10 µg), enrofloxacin (ENR: 10 µg), cefixime (CFM: 5 µg) and tetracycline (TET: 30 µg). The zone diameters were measured (to the nearest millimetre) and interpreted as intermediate (I), susceptible (S) or resistant (R) according to CLSI protocol (Wayne 2012a); intermediate strains were considered susceptible. Based on the definition proposed by an international expert, the MDR phenotype was resistant to three or more antimicrobial classes (Magiorakos et al. 2012). E. coli, ATCC 25922 (sensitive to all these drugs), recommended by CLSI, was used as a quality control. The specified range of quality control result was published in M100-S22 (Wayne 2012b).
DNA extraction
A single colony of overnight TSB culture was suspended in 100 µL of distilled water and exposed to boiling for 10 min at 100 °C. After a 13 min freeze, the frozen cell pellets were centrifuged at 14 000 rpm for 10 min (Dehkordi et al. 2014) and the supernatant, containing bacterial DNA, was subjected to PCR analysis.
Polymerase chain reaction detection of virulence factors and class 1 integron in Shiga toxin-producing Escherichia coli strains
Polymerase chain reaction assays were used to detect the presence of the following virulence genes coding regions including stx1, stx2 and eae. To detect class 1 integron in confirmed STEC isolates, a PCR protocol was employed. Preparation of the DNA samples was done as described in previously published paper (Dehkordi et al. 2014). Primer sequences, sizes of PCR products and PCR conditions are shown in Table 1. DNA from E. coli O157:H7 EDL933 strain and ATCC 25922 strains were used as positive and negative controls, respectively. The amplified DNA products were separated by 1.5% agarose gel electrophoresis (Sigma-Aldrich, St. Louis, MO, United States). The gels were stained with ethidium bromide (Merek, Germany). Visualisation of amplified products was done by ultraviolet (UV) illumination and photographed using a Kodak camera system (Gel Logic 200).
TABLE 1.
Primers and polymerase chain reaction conditions used in this study.
| Gene | Primer sequence | Size of product (bp) | PCR programme | PCR volume (25 μL) | Reference |
|---|---|---|---|---|---|
| stx1 | F: CTT CGG TAT CCT ATT CCC GG R: GGA TGC ATC TCT GGT CAT TG |
484 | 25 cycles of 30 s at 94 °C 45 s at 50 °C 90 s at 70 °C 10 min at 70 °C |
2.5 μL PCR buffer 10X 1.25 μL MgCl2 0.5 μLdNTP 1 μL of each primers F & R 0.25 μLTaq DNA polymerase 1 μL DNA template |
Tahamtan et al. (2010) |
| stx2 | F: CCA TGA CAA CGG ACA GCA GTT R: CCT GTC AAC TGA GCA GCA CTT TG |
779 | 25 cycles of 30 s at 94 °C 45 s at 50 °C 90 s at 70 °C 10 min at 70 °C |
2.5 μL PCR buffer 10X 1.25 μL MgCl2 0.5 μLdNTP 1 μL of each primers F & R 0.25 μLTaq DNA polymerase 1 μL DNA template |
Tahamtan et al. (2010) |
| eae | F: AAG CGA CTG AGG TCA CT R: ACG CTG CTC ACT AGA TGT |
384 | 25 cycles of 30 s at 94 °C 45 s at 50 °C 90 s at 70 °C 10 min at 70 °C |
2.5 μL PCR buffer 10X 1.25 μL MgCl2 0.5 μLdNTP 1 μL of each primers F & R 0.25 μLTaq DNA polymerase 1 μL DNA template |
Vasconcellos et al. (2012) |
| IntI | F: TGCGGGTYAARGATBTKGATTT* R: CARCACATGCGTRTARAT |
491 | 30 s at 94 °C, 35 s at 57 °C 25 cycles of 1 min at 70 °C 10 min at 72 °C |
2.5 μL PCR buffer X 10 1.25 μL MgCl2 1 μLdNTP 1 μL of each primers F & R 0.25 μLTaq DNA polymerase 1 μl DNA template |
Tahamtan et al. (2014) |
Note: B = C or G or T; K = G or T; R = A or G; Y = C or T*.
PCR, polymerase chain reaction; DNA, deoxyribonucleic acid; Taq, thermus aquaticus; stx, isolates carrying stx1 and/or stx2 genes; eae, isolates carrying eae gene; pb, base pair; IntI, encoded integrases.
Statistical analysis
The chi-square (χ2) test and Fisher’s exact test were used to assess whether integron-positive strains were significantly more resistant than integron-negative strains for each of the tested antibiotics. A p value < 0.05 was considered statistically significant. Statistical calculations were made using GraphPad Prism for Windows version 5 (GraphPad Software, San Diego, CA).
Results
Isolation and characterisation of Shiga toxin-producing Escherichia coli in calves
From 540 diarrhoeic calves, 312 samples (57.7%) were positive for E. coli. Shiga toxin–producing Escherichia coli strains were isolated from 71 (22.7%) out of the 312 samples, which possess stx1 and/or stx2. Twelve (3.57%) isolates were classified as E. coli O157:H7 and 59 (31.19%) as non-O157.
Characterisation of virulence genes
Of 312 E. coli strains, 71 isolates (22.7%) were identified as STEC. The virulence genes stx2, stx1 and eae were detected at 76%, 46.4% and 53.5% in STEC isolates, respectively. Of isolates that were not characterised as STEC, 101 (32.3%) were positive for eae gene (Figures 1 and 2). These findings are summarised in Table 2. Out of 59 non-O157 strains (PNU6, PNU11, PNU12 and PNU16) in the diarrhoeic calves, four were positive for both the stx1 and stx2 genes and three non-O157 harboured all of the stx1, stx2 and eae genes.
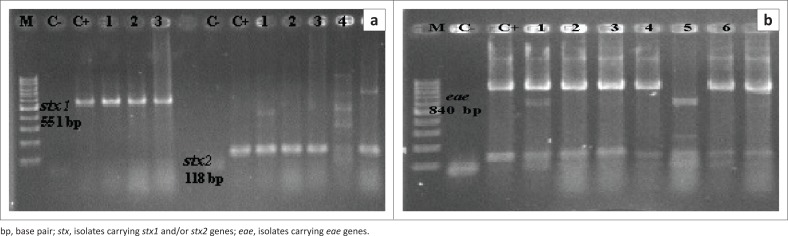
FIGURE 1.
Agarose gels electerophoresis of Shiga toxin-producing Escherichia coli isolates. (a) Polymerase chain reaction amplification of the stx1 (551 bp) and stx2 (118 bp) genes. Lanes 1–3, stx1; lanes 1–3 and 5, stx2 and (b) polymerase chain reaction amplification of the eae gene (840 bp) (lanes 1–4, 6 and 7). Lane M, 100 bp molecular size markers; Lanes C- and C+, negative and positive control.
FIGURE 2.
Frequency of occurrence of tested virulence genes in 71 STEC strains.
TABLE 2.
Distribution of virulence genes in Shiga toxin-producing Escherichia coli strains.
| Pathotype | Serogroup | Positive sample |
Number of isolates carrying specific genes |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % |
stx1 |
stx2 |
eae |
stx2/stx1 |
stx1/eae |
stx2/eae |
stx1/stx2/eae |
|||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | ||||
| STEC | Non-O157 | 59 | 100.0 | 21 | 36 | 41 | 70 | 26 | 44.0 | 3 | 5.0 | 10 | 16.9 | 13 | 22.0 | 3 | 5.0 |
| O157 | 12 | - | 12 | - | 12 | - | 12 | - | 0 | - | 0 | - | 0 | - | 12 | - | |
| Total STEC | 71 | 100.0 | 33 | 46 | 53 | 75 | 38 | 54 | 3 | 4.2 | 10 | 14.0 | 13 | 18.3 | 15 | 21.1 | |
| Non-STEC | 241 | - | - | - | - | - | 63 | - | - | - | - | - | - | - | - | - | |
| Total | 312 | 100.0 | 33 | 11 | 53 | 17.0 | 101 | 32 | 3 | 0.9 | 10 | 3.2 | 13 | 4.1 | 15 | 4.8 | |
Overall: STEC, 71 (22.7%); Non-STEC, 241 (77.3%).
Overall: Non-O157, 52 (16.6%); O157, 12 (3.8%).
STEC, Shiga toxin-producing Escherichia coli; No., number; stx, isolates carrying stx1 and/or stx2 genes; eae, isolates carrying eae gene.
Note: The eae gene produces a 94-kDa outer membrane protein called intimin.
Antibiotic susceptibility
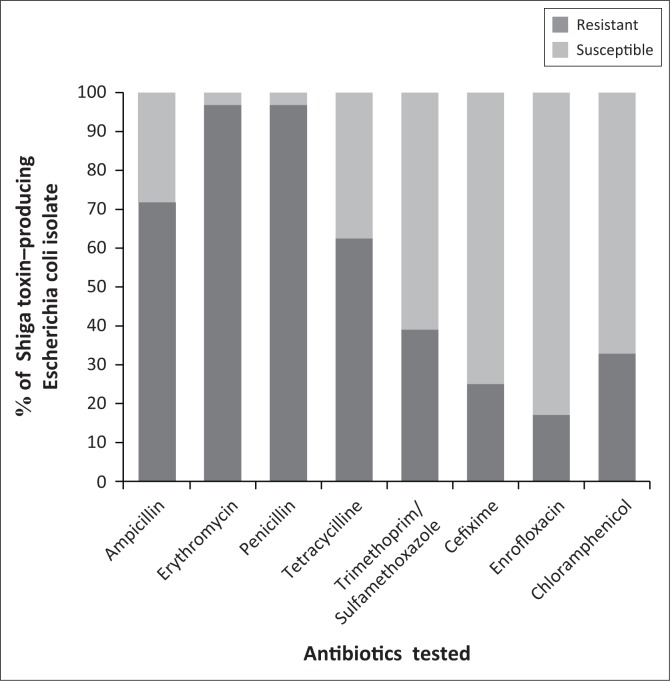
The antimicrobial susceptibility of 52 non-O157 and 12 O157 STEC isolates was determined by the disk diffusion method. The resistance patterns of the E. coli O157 strains were to penicillin and ampicillin (91% – 8%), followed by tetracycline, erythromycin and cefixime (66% – 25%). Nine (75%) of the 12 O157 strains exhibited multidrug resistance (MDR, resistant to ≥ 3 antimicrobial classes). The most common MDR phenotypes were AM-E-P-TET, which accounted for 15% of the 12 O157 strains. All of the examined non-O157 strains showed resistance to trimethoprim/sulfamethoxazole. The resistance patterns of all non-O157 strains to tested antibiotics were as follows: erythromycin (98%), penicillin (91%), ampicillin (73%), tetracycline (65%), chloramphenicol (40%), cefixime (25%) and enrofloxacin (21%). Forty-eight (92%) of the 52 non-O157 strains displayed multidrug resistance. The most frequently observed MDR profiles AM-C-E-P-TET-ENR–SXT (33% of the 52 non-O157 strains) were associated with 10 of these isolates (PNU4, PNU29, PNU31, PNU33, PNU38, PNU41, PNU49, PNU50, PNU51, PNU52). The resistance patterns of 64 STEC isolates are shown in Table 3 and Figure 3.
TABLE 3.
Antibiotic resistance pattern in Shiga toxin–producing Escherichia coli strains.
| STEC - Serogroup | No. positive |
AM10 |
C30 |
CFM5 |
E25 |
ENR10 |
P10 |
SXT30 |
TET30 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | % | Sample | % | Sample | % | Sample | % | Sample | % | Sample | % | Sample | % | Sample | % | Sample | % | |
| Non-O157 | 52 | 100.0 | 38 | 73.0 | 21 | 40.0 | 13 | 25.0 | 51 | 98.0 | 11 | 21.1 | 51 | 98.0 | 52 | 100.0 | 34 | 65.3 |
| O157 | 12 | 18.7 | 10 | 83.0 | 0 | 3 | 25.0 | 8 | 66.6 | 0 | - | 11 | 91.6 | 0 | 8 | 66.6 | ||
| Total STEC | 64 | 100.0 | 48 | 75.0 | 21 | 32.8 | 16 | 25.0 | 59 | 92.1 | 11 | 17.1 | 62 | 96.8 | 25 | 39.0 | 42 | 65.6 |
AM10, ampicillin (10 µg/disk); TET30, tetracycline (30 µg/disk); E25, erythromycin (25 µg/disk); ENR10, enrofloxacin (10 µg/disk); SXT30, trimethoprim/sulfamethoxazole (30 µg/disk); C30, chloramphenicol (30 µg/disk); P10, penicillin (10 µg/disk); CFM5, cefixime (5 µg/disk); STEC, Shiga toxin-producing Escherichia coli; No., number.
FIGURE 3.
Antimicrobial susceptibility patterns in 71 Shiga toxin-producing Escherichia coli strains.
Integrons
Class 1 integrons were detected among 23 (36%) of the STEC isolates (Figure 4 and Table 4). Integron-positive strains were significantly more resistant to enrofloxacin, trimethoprim/sulfamethoxazole and tetracycline than integron-negative strains (p < 0.05). Nevertheless, resistance to ampicillin, erythromycin, penicillin, cefixime and chloramphenicol could not be directly related to the presence of integrons (Table 5). All of the integron-positive strains displayed multidrug resistance. The most prevalent MDR phenotypes in integron-positive strains were AM-CFM-E-P-TET (26% of the 52 non-O157 strains).
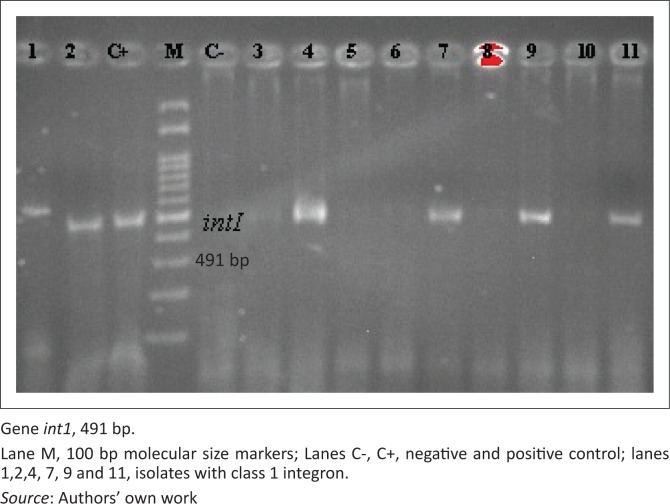
FIGURE 4.
Polymerase chain reaction amplicons of Shiga toxin-producing Escherichia coli integrons. Polymerase chain reaction amplification of the class 1 integron, integrase. bp, base pair; int1, integrase gene.
TABLE 4.
Overview of the integron-positive Shiga toxin–producing Escherichia coli strains.
| Strain | Serogroup | Virulence profile stx1/2 gene | Integron int gene | Antibiotic resistance profile |
MDR | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | C | CFM | E | ENR | P | SXT | TET | |||||
| PNU1 | Non-157 | 1 | + | R | S | S | R | S | R | R | R | + |
| PNU2 | Non-157 | 2 | + | S | S | S | R | S | R | R | R | + |
| PNU3 | Non-157 | 2 | + | R | R | S | R | R | S | R | R | + |
| PNU4 | Non-157 | 2 | + | R | R | S | R | R | R | R | R | + |
| PNU5 | Non-157 | 2 | + | R | S | S | R | S | R | S | R | + |
| PNU7 | Non-157 | 1 | + | R | S | R | R | S | R | S | R | + |
| PNU10 | Non-157 | 1 | + | R | S | R | R | S | R | S | R | + |
| PNU20 | O157 | 1, 2 | + | R | S | R | R | S | R | S | R | + |
| PNU21 | O157 | 1, 2 | + | R | S | R | R | S | R | S | R | + |
| PNU24 | O157 | 1, 2 | + | R | S | S | R | S | R | S | R | + |
| PNU26 | O157 | 1, 2 | + | S | S | S | R | S | R | S | S | - |
| PNU30 | Non-157 | 2 | + | R | R | S | R | R | R | R | R | + |
| PNU31 | Non-157 | 2 | + | R | R | S | R | R | R | R | R | + |
| PNU33 | Non-157 | 1 | + | R | R | S | R | R | R | R | R | + |
| PNU34 | Non-157 | 2 | + | S | R | S | R | S | R | R | R | + |
| PNU35 | Non-157 | 2 | + | R | S | R | R | S | R | S | R | + |
| PNU39 | Non-157 | 2 | + | S | R | S | R | S | R | R | R | + |
| PNU40 | Non-157 | 1 | + | R | S | S | R | R | R | R | R | + |
| PNU50 | Non-157 | 1 | + | R | R | S | R | R | R | R | R | + |
| PNU51 | Non-157 | 2 | + | R | R | R | R | S | R | R | R | + |
| PNU58 | Non-157 | 2 | + | R | S | S | R | S | R | R | R | + |
| PNU61 | Non-157 | 2 | + | R | R | S | R | S | R | R | R | + |
| PNU62 | Non-157 | 2 | + | R | S | S | R | S | R | R | R | + |
Note: Antibiotic resistance profile was determined for eight antibiotics: ampicillin (AM), tetracycline (TET), erythromycin (E), enrofloxacin (ENR), trimethoprim/sulfamethoxazole (SXT), chloramphenicol (C), penicillin (P) and cefixime (CFM).
stx, isolates carrying stx1 and/or stx2 genes; MDR, multidrug-resistant isolates; S, antibiotic-susceptible isolates; R, antibiotic-resistant isolates.
+, positive for int gene or MDR.
TABLE 5.
Comparison of the resistances between integron-positive and integron-negative strains was done using the p-values listed in the table.
| Antibiotic | Resistance int-positive isolates |
Resistance int-negative isolates |
Resistance of total isolates |
Association with integron | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||
| Ampicillin | 19 | 29.6 | 28 | 42.2 | 47 | 71.8 | 0.2523 |
| Erythromycin | 23 | 35.9 | 39 | 60.9 | 62 | 96.8 | 0.5322 |
| Penicillin | 22 | 34.3 | 40 | 62.5 | 62 | 96.8 | 1.0000 |
| Tetracycline | 21 | 32.8 | 20 | 29.6 | 41 | 62.5 | 0.0009* |
| Trimethoprim/sulfamethoxazole | 15 | 23.4 | 10 | 15.6 | 15 | 39.00 | 0.0032* |
| Cefixime | 7 | 10.9 | 9 | 14.1 | 16 | 25.00 | 0.6521 |
| Enrofloxacin | 7 | 10.9 | 4 | 6.2 | 11 | 17.1 | 0.0456* |
| Chloramphenicol | 10 | 15.6 | 11 | 17.1 | 21 | 32.8 | 0.2780 |
Note: p values of 0.05 were considered to be significant.
int-positive, integron-positive in PCR assay; int-negative, integron-negative in PCR assay.
int, integron; No., number.
, Correlation is significant at the 0.05 level.
Discussion
Antibiotic resistance developed in STEC isolates from humans and animals (Van Meervenne et al. 2013). Integrons, which are known to be associated with many antimicrobial resistance genes, were suspected to serve as pools of antimicrobial resistance genes worldwide (El-Sokkary & Abdelmegeed 2015). Class 1 integrons are commonly found in gram-negative pathogens (Maguire et al. 2001).
In this study, the presence of major virulence factors and resistance to antimicrobials belonging to classes generally utilised in Iran was investigated in zoonotic STEC isolates from calves with diarrhoea. Owing to the close contact of humans with animals, the presence of virulence and antimicrobial resistance genes in E. coli strains harboured by animals leads to public health concerns (Torkan et al. 2016). Escherichia coli, which has been implicated as an aetiological factor of calf diarrhoea, harbours many virulence genes that enable it to cause disease in a particular host (Nagarjuna et al. 2015). In the present study, among 312 E. coli strains from diarrhoeic calves, 71 (22.7%) were STEC. The results are in agreement with those of Dastmalchi et al. (2012), who screened 51 E. coli isolates from diarrhoeic calves in the Urmia region, which is located in west Azerbaijan Province, Iran, and illustrated that 19.6% of isolates were stx positive. Most epidemiological studies in diarrhoeic calves in Iran have disclosed that the prevalence of STEC infection ranges between 6.4% and 34.5% (Pourtaghi, Dahpahlavan & Momtaz 2013; Shahrani et al. 2014). These discrepancies can be attributed to the small sample size and geographical differences. In other words, STEC prevalence in calves may be influenced by environmental factors (Dastmalchi et al. 2012). Higher prevalence of the stx2 gene (54 isolates) compared to the stx1 gene (33 isolates) in this study corroborates the findings of previous reports in Iran (Dastmalchi et al. 2012; Tahamtan, Hayati & Namavari 2010). However, these results contrast with other reports that have shown that most STEC from diarrhoeic calves only produce stx1, whereas stx2-positive strains are the dominant types in healthy calves (Nguyen, Vo & Vu-Khac 2011). The differences in these findings suggest that stx2 may be associated with a majority of E. coli isolates from diarrhoeic calves in Iran. Shiga toxin producing E. coli infection, which is associated with diarrhoea in calves, may result in severe diseases in humans such as HUS and HC (Bastos et al. 2006). The diarrhoeal phase of diseases associated with STEC is usually self-limiting, and the role of early antimicrobial treatment in the prevention of HUS is still regarded as controversial (Shahrani et al. 2014). Current recommendations and the available data suggest that not only do antibiotic exposure increase the risk of HUS in children via inducing expression of stx through replication of temperate bacteriophages carrying stx-encoding genes (Ochoa et al. 2007), it turns out to have another perilous effect on the frequency of STEC antimicrobial resistance (Shahrani et al. 2014), which could result in an increase of frequency of STEC and perhaps greater shedding. Resistance could contribute to greater contamination of animal food products with STEC (Torkan et al. 2016). Several reports have documented that a significant increase of antimicrobial resistance in STEC strains isolated from animals and humans has acquired antibiotic resistance genes almost 20 years ago (Zhao et al. 2001). In STEC strains, class 1 integrons are strongly associated with multidrug resistance (Colello et al. 2015). Previous studies have reported the occurrence and prevalence of class 1 integrons to be ranging from 2.7% to 41.0% among STEC isolates in Germany (Askar et al. 2011), Argentina (Colello et al. 2015), Belgium (Van Meervenne et al. 2013), North America (Nagachinta & Chen 2009), Brazil (Cergole-Novella et al. 2011) and US (Singh et al. 2005; Zhao et al. 2001). Class 1 integrons appear to be common in the endemic STEC strains. In the present study, class 1 integron was identified in 23 (36%) out of 71 STEC isolates. Our data revealed low distribution of class 1 integrons among STEC isolates from calves with diarrhoea in the south of Iran compared with a similar study in northern Iran in 2014 for which the authors found a higher percentage (53%) of the strains containing integron class 1 (Bakhshi et al. 2014). The various percentages of class 1 integrons in different parts of the world could be attributed to the characteristics of the analysed collection and differences in the prevalence of antibiotic consumption in each country (Kargar et al. 2014). In general, exposure to antibiotics, heavy metals or biocides and a high multiplicity of other different environmental factors are among the main reasons for an increase of cells containing integrons (Baquero, Martínez & Cantón 2008).
All integron-positive strains examined in this study were resistant to at least three different antibiotics (MDR). Similarly, high percentages of MDR phenotypes among integron-positive STEC strains have been reported in Argentina (Nagachinta & Chen 2009) and Iran (Bakhshi, Najibi & Sepehri-Seresht 2014). However, other authors (Colello et al. 2015; Van Meervenne et al. 2013) have reported a lower rate (less than 90%) of STEC in diarrhoeic calves. The highest resistances among the integron-positive strains were found to enrofloxacin (17%), trimethoprim/sulfamethoxazole (39%) and tetracycline (62%). The integron-positive strains were significantly more resistant to these antibiotics than the integron-negative strains. The resistance to enrofloxacin (ENR), trimethoprim/sulfamethoxazole (SXT) and tetracycline (TET) is related to the presence of the integron. The significant association between resistance to fluoroquinolones, tetracycline, trimethoprim and sulfonamides (ENR, TET, SXT) tested and integron existence could be explained because of the fact that many fluoroquinolone, tetracycline, trimethoprim and sulfonamide resistant genes have been reported within integron structures, including gyrA, gyrB, qnr, tetA, tetB, tetC, tetD, sul1, sul2, sul3 and dfrA1 (Kaplan et al. 2013; Wang et al. 2010).
Conclusion
We report the presence of class 1 integrons in the most familiar STEC strains from diarrhoeic calves. Results imply that stx2, stx1 and eae putative virulence gene, the IntI integrase gene and resistance to erythromycin, penicillin, ampicillin, tetracycline and trimethoprim/sulfamethoxazole were the most commonly detected characteristics of the STEC strains isolated from diarrhoeic calves in southern Iran. Our investigation demonstrated that calves are possible reservoirs of STEC strains and developed resistance to multiple classes of antimicrobials. Emerging data suggest an association between MDR and integrons which may play a significant role in the dissemination of resistance genes. Therefore, it is advised to stop routine antimicrobial treatment and conduct further molecular studies to detect other antimicrobial resistance and virulent genes in STEC isolates obtained in this study.
Acknowledgements
Competing interests
The authors declare that they have no financial or personal relationships which may have inappropriately influenced them in writing this paper.
Authors’ contributions
M.K. contributed to culture, DNA extraction, PCR techniques, analysis of results, statistical analysis and writing of the manuscript. A.G. carried out writing and revision of the manuscript. Both authors read and approved the final manuscript.
Footnotes
How to cite this article: Kohansal, M. & Ghanbari Asad, A., 2018, ‘Molecular analysis of Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains isolated from calves’, Onderstepoort Journal of Veterinary Research 85(1), a1621. https://doi.org/10.4102/ojvr.v85i1.1621
References
- Askar M., Faber M.S., Frank C., Bernard H., Gilsdorf A., Fruth A. et al. , 2011, ‘Update on the ongoing outbreak of haemolytic uraemic syndrome due to Shiga toxin-producing Escherichia coli (STEC) serotype O104, Germany’, Eurosurveillance 16(22), 19883. [DOI] [PubMed] [Google Scholar]
- Bakhshi B., Najibi S. & Sepehri-Seresht S, 2014, ‘Molecular characterization of enterohemorrhagic Escherichia coli isolates from cattle’, Journal of Veterinary Medical Science 76(9), 1195–1199. https://doi.org/10.1292/jvms.13-0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F., Martínez J.L. & Cantón R, 2008, ‘Antibiotics and antibiotic resistance in water environments’, Current Opinion in Biotechnology 19(3), 260–265. https://doi.org/10.1016/j.copbio.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Bastos F.C., Vaz T.M.I., Irino K. & Guth B.E.C, 2006, ‘Phenotypic characteristics, virulence profile and genetic relatedness of O157 Shiga toxin-producing Escherichia coli isolated in Brazil and other Latin American countries’, FEMS Microbiology Letters 265(1), 89–97. https://doi.org/10.1111/j.1574-6968.2006.00472.x [DOI] [PubMed] [Google Scholar]
- Cergole-Novella M.C., Pignatari A.C.C., Castanheira M. & Guth B.E.C, 2011, ‘Molecular typing of antimicrobial-resistant Shiga-toxin-producing Escherichia coli strains (STEC) in Brazil’, Research in Microbiology 162(2), 117–123. https://doi.org/10.1016/j.resmic.2010.09.022 [DOI] [PubMed] [Google Scholar]
- Colello R., Etcheverría A.I., Conza J.A.D., Gutkind G.O. & Padola N.L, 2015, ‘Antibiotic resistance and integrons in Shiga toxin-producing Escherichia coli (STEC)’, Brazilian Journal of Microbiology 46(1), 1–5. https://doi.org/10.1590/S1517-838246120130698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastmalchi S.H. & Ayremlou N, 2012, ‘Characterization of Shiga toxin-producing Escherichia coli (STEC) in feces of healthy and diarrheic calves in Urmia region, Iran’, Iranian Journal of Microbiology 4(2), 63. [PMC free article] [PubMed] [Google Scholar]
- Dehkordi F.S., Yazdani F., Mozafari J. & Valizadeh Y, 2014, ‘Virulence factors, serogroups and antimicrobial resistance properties of Escherichia coli strains in fermented dairy products’, BMC Research Notes 7(1), 217 https://doi.org/10.1186/1756-0500-7-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Verdier K., Nyman A., Greko C. & Bengtsson B, 2012, ‘Antimicrobial resistance and virulence factors in Escherichia coli from Swedish dairy calves’, Acta Veterinaria Scandinavica 54(1), 2 https://doi.org/10.1186/1751-0147-54-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekhari N., Bakhshi B., Pourshafie M.R., Zarbakhsh B., Rahbar M., Hajia M. et al. , 2013, ‘Genetic diversity of Shigella spp. and their integron content’, Foodborne Pathogens and Disease 10(3), 237–242. https://doi.org/10.1089/fpd.2012.1250 [DOI] [PubMed] [Google Scholar]
- El-Sokkary M.M.A. & Abdelmegeed E.S, 2015, ‘Characterisation of class 1 integron among Escherichia coli isolated from Mansoura University hospitals in Egypt’, Advances in Microbiology 5(4), 269 https://doi.org/10.4236/aim.2015.54025 [Google Scholar]
- Gonzalez G., Sossa K., Bello H., Dominguez M., Mella S. & Zemelman R, 1998, ‘Presence of integrons in isolates of different biotypes of Acinetobacterbaumannii from Chilean hospitals’, FEMS Microbiology Letters 161(1), 125–128. https://doi.org/10.1111/j.1574-6968.1998.tb12937.x [DOI] [PubMed] [Google Scholar]
- GraphPad , 2005, Prism version 5, GraphPad Software, San Diego, CA. [Google Scholar]
- Hamada K., Oshima K. & Tsuji H, 2003, ‘Drug resistance genes encoded in integrons and in extra-integrons: Their distribution and lateral transfer among pathogenic Enterobacteriaceae including enterohemorrhagic Escherichia coli and Salmonella enterica serovars typhimurium and infantis’, Japanese Journal of Infectious Diseases 56(3), 123–126. [PubMed] [Google Scholar]
- Kaplan E., Offek M., Jurkevitch E. & Cytryn E, 2013, ‘Characterization of fluoroquinolone resistance and qnr diversity in Enterobacteriaceae from municipal biosolids’, Frontiers in Microbiology 4, 144 https://doi.org/10.3389/fmicb.2013.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargar M., Mohammadalipour Z., Doosti A., Lorzadeh S. & Japoni-Nejad A, 2014, ‘High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in southwest of Iran’, Osong Public Health and Research Perspectives 5(4), 193–198. https://doi.org/10.1016/j.phrp.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G. et al. , 2012, ‘Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance’, Clinical Microbiology and Infection 18(3), 268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Maguire A.J., Brown D.F., Gray J.J. & Desselberger U, 2001, ‘Rapid screening technique for class 1 integrons inenterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology’, Antimicrobial Agents and Chemotherapy 45(4), 1022–1029. https://doi.org/10.1128/AAC.45.4.1022-1029.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Freijo P., Fluit A.C., Schmitz F.J., Grek V.S., Verhoef J. & Jones M.E, 1998, ‘Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds’, The Journal of Antimicrobial Chemotherapy 42(6), 689–696. [DOI] [PubMed] [Google Scholar]
- Martinez-Freijo P., Fluit A.C., Schmitz F.J., Verhoef J. & Jones M.E, 1999, ‘Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe’, Antimicrobial Agents and Chemotherapy 43(3), 686–689. https://doi.org/10.1093/jac/42.6.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro M.A., Bülte M., Trumpf T., Aleksić S., Reuter G., Bulling E. et al. , 1990, ‘Detection and characterization of fecal verotoxin-producing Escherichia coli from healthy cattle’, Journal of Clinical Microbiology 28(6), 1417–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura C.D., Ludovico M., Valadares G.F., Gatti M.S.V. & Leite D.S, 2012, ‘Detection of virulence genes in Escherichia coli strains isolated from diarrheic and healthy feces of dairy calves in Brazil’, Arquivos do InstitutoBiológico 79(2), 273–276. https://doi.org/10.1590/S1808-16572012000200016 [Google Scholar]
- Nagachinta S. & Chen J, 2009, ‘Integron-mediated antibiotic resistance in Shiga toxin–producing Escherichia coli’, Journal of Food Protection 72(1), 21–27. https://doi.org/10.4315/0362-028X-72.1.21 [DOI] [PubMed] [Google Scholar]
- Nagarjuna D., Mittal G., Dhanda R.S., Verma P.K., Gaind R. & Yadav M, 2015, ‘Faecal Escherichia coli isolates show potential to cause endogenous infection in patients admitted to the ICU in a tertiary care hospital’, New Microbes and New Infections 7, 57–66. https://doi.org/10.1016/j.nmni.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najibi S., Bakhshi B., Fallahzad S., Pourshafie M.R., Katouli M., Sattari M. et al. , 2012, ‘Distribution of class 1 integrons among enteropathogenic Escherichia coli’, Canadian Journal of Microbiology 58(5), 637–643. https://doi.org/10.1139/w2012-035 [DOI] [PubMed] [Google Scholar]
- Nguyen T.D., Vo T.T. & Vu-Khac H, 2011, ‘Virulence factors in Escherichia coli isolated from calves with diarrhea in Vietnam’, Journal of Veterinary Science 12(2), 159–164. https://doi.org/10.4142/jvs.2011.12.2.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa T.J., Chen J., Walker C.M., Gonzales E. & Cleary T.G, 2007, ‘Rifaximin does not induce toxin production or phage-mediated lysis of Shiga toxin-producing Escherichia coli’, Antimicrobial Agents and Chemotherapy 51(8), 2837–2841. https://doi.org/10.1128/AAC.01397-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourtaghi H., Dahpahlavan V. & Momtaz H, 2013, ‘Virulence genes in Escherichia coli isolated from calves with diarrhoea in Iran’, Comparative Clinical Pathology 22(3), 513–515. https://doi.org/10.1007/s00580-012-1442-5 [Google Scholar]
- Pradel N., Bertin Y., Martin C. & Livrelli V, 2008, ‘Molecular analysis of shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France’, Applied and Environmental Microbiology 74(7), 2118–2128. https://doi.org/10.1128/AEM.02688-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigobelo E.C., Gamez H.J., Marin J.M., Macedo C., Ambrosin J.A. & Ávila F.A.D, 2006, ‘Virulence factors of Escherichia coli isolated from diarrheic calves’, Arquivo Brasileiro de Medicina Veterinária e Zootecnia 58(3), 305–310. https://doi.org/10.1590/S0102-09352006000300003 [Google Scholar]
- Schroeder C.M., Zhao C., DebRoy C., Torcolini J., Zhao S., White D.G. et al. , 2002, ‘Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food’, Applied and Environmental Microbiology 68(2), 576–581. https://doi.org/10.1128/AEM.68.2.576-581.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams Z., Tahamtan Y., Pourbakhsh A., Hosseiny M.H., Kargar M. & Hayati M, 2012, ‘Detection of enterotoxigenic K99 (F5) and F41 from fecal sample of calves by molecular and serological methods’, Comparative Clinical Pathology 21(4), 475–478. https://doi.org/10.1007/s00580-010-1122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrani M., Dehkordi F.S. & Momtaz H, 2014, ‘Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran’, Biological Research 47(1), 28 https://doi.org/10.1186/0717-6287-47–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Schroeder C.M., Meng J., White D.G., McDermott P.F., Wagner D.D. et al. , 2005, ‘Identification of antimicrobial resistance and class 1 integrons in Shiga toxin-producing Escherichia coli recovered from humans and food animals’, Journal of Antimicrobial Chemotherapy 56(1), 216–219. https://doi.org/10.1093/jac/dki161 [DOI] [PubMed] [Google Scholar]
- Tahamtan Y., AsadiFozi M., Hosseini M. & Mostafaee T, 2014, ‘Molecular characterization of class 1 integrons and antibiotic resistance to E. coli from cattle, sheep and goats’, Online Journal of Veterinary Research 18(9), 701–709. [Google Scholar]
- Tahamtan Y., Hayati M. & Namavari M.M, 2010, ‘Prevalence and distribution of the stx1, stx2 genes in Shiga toxin producing E. coli (STEC) isolates from cattle’, Iranian Journal of Microbiology 2(1), 8. [PMC free article] [PubMed] [Google Scholar]
- Thomas K.M., McCann M.S., Collery M.M., Logan A., Whyte P., McDowell D.A. et al. , 2012, ‘Tracking verocytotoxigenic Escherichia coli O157, O26, O111, O103 and O145 in Irish cattle’, International Journal of Food Microbiology 153(3), 288–296. https://doi.org/10.1016/j.ijfoodmicro.2011.11.012 [DOI] [PubMed] [Google Scholar]
- Torkan S., Bahadoranian M.A., Khamesipour F. & Anyanwu M.U, 2016, ‘Detection of virulence and antimicrobial resistance genes in Escherichia coli isolates from diarrhoiec dogs in Iran’, Archivos de Medicina Veterinaria 48(2), 181–190. https://doi.org/10.4067/S0301-732X2016000200008 [Google Scholar]
- Van Meervenne E., Boon N., Verstraete K., Devlieghere F., De Reu K., Herman L. et al. , 2013, ‘Integron characterization and typing of Shiga toxin-producing Escherichia coli isolates in Belgium’, Journal of Medical Microbiology 62(5), 712–719. https://doi.org/10.1099/jmm.0.048934-0 [DOI] [PubMed] [Google Scholar]
- Vasconcellos H.L.F., Scaramuzzi K., Nascimento I.P., Ferreira J.M.D.C. Jr., Abe C.M., Piazza R.M. et al. , 2012, ‘Generation of recombinant bacillus Calmette–Guérin and Mycobacterium smegmatis expressing BfpA and intimin as vaccine vectors against enteropathogenic Escherichia coli’, Vaccine 30(41), 5999–6005. https://doi.org/10.1016/j.vaccine.2012.05.083 [DOI] [PubMed] [Google Scholar]
- Wang Y.C., Chan J.P.W., Yeh K.S., Chang C.C., Hsuan S.L., Hsieh Y.M. et al. , 2010, ‘Molecular characterization of enrofloxacin resistant Actinobacillus pleuropneumoniae isolates’, Veterinary Microbiology 142(3–4), 309–312. https://doi.org/10.1016/j.vetmic.2009.09.067 [DOI] [PubMed] [Google Scholar]
- Wayne P.A, 2012a, Clinical and laboratory standards: Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobicall, Approved Standard M07-A9, 9th edn, Clinical and Laboratory Standards Institute, PA, USA. [Google Scholar]
- Wayne P.A, 2012b, Clinical and laboratory standards: Performance standards for antimicrobial disk susceptibility tests, Approved Standard M02-A11, 11th edn, Clinical and Laboratory Standards Institute, PA, USA. [Google Scholar]
- White P.A., McIver C.J. & Rawlinson W.D, 2001, ‘Integrons and gene cassettes in the Enterobacteriaceae’, Antimicrobial Agents and Chemotherapy 45(9), 2658–2661. https://doi.org/10.1128/AAC.45.9.2658-2661.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., White D.G., Ge B., Ayers S., Friedman S., English L. et al. , 2001, ‘Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates’, Applied and Environmental Microbiology 67(4), 1558–1564. https://doi.org/10.1128/AEM.67.4.1558-1564.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]